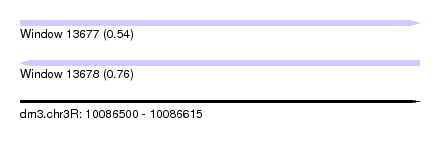
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 10,086,500 – 10,086,615 |
| Length | 115 |
| Max. P | 0.758813 |

| Location | 10,086,500 – 10,086,615 |
|---|---|
| Length | 115 |
| Sequences | 10 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 67.57 |
| Shannon entropy | 0.70679 |
| G+C content | 0.67217 |
| Mean single sequence MFE | -52.11 |
| Consensus MFE | -19.46 |
| Energy contribution | -19.95 |
| Covariance contribution | 0.49 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.541101 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
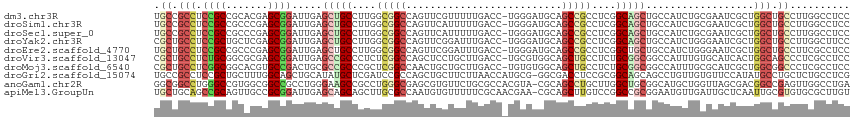
>dm3.chr3R 10086500 115 + 27905053 UGCCGCCUCCGCCGCACGAGCGGAUUGAGCUGCCUUGGCGGCCAGUUCGUUUUUGACC-UGGGAUGCAGCCGCCUCGGCAGCUGCCAUCUGCGAAUCGCUGGCUGCCUUGGCCUCC ....((.((..((((....))))...)))).(((..((((((((((((((........-..(((((((((.((....)).)))).)))))))))...))))))))))..))).... ( -53.30, z-score = -1.71, R) >droSim1.chr3R 16152480 115 - 27517382 UGCCGCCUCCGCCGCCCGAGCGGAUUGAGCUGCCUUGGCGGCCAGUUCAUUUUUGACC-UGGGAUGCAGCCGCCUCGGCAGCUGCCAUCUGCGAAUCGCUGGCUGCCUUGGCCUCC ....(((...((.((((((((((((.((((((((..(((((((((.(((....))).)-))(....).))))))..))))))).).))))))...)))..))).))...))).... ( -52.70, z-score = -1.76, R) >droSec1.super_0 9166967 115 - 21120651 UGCCGCCUCCGCCGCCCGAGCGGAUUGAGCUGCCUUGGCGGCCAGUUCAUUUUUGACC-UGGGAUGCAGCCGCCUCGGCAGCUGCCAUCUGCGAAUCGCUGGCUGCCUUGGCCUCC ....(((...((.((((((((((((.((((((((..(((((((((.(((....))).)-))(....).))))))..))))))).).))))))...)))..))).))...))).... ( -52.70, z-score = -1.76, R) >droYak2.chr3R 12470708 115 - 28832112 CGCUGCCUCCGCUGCUCGAGCGGAUUGAGCUGCCUUGGCGGCCAGUUCGGAUUUGACC-UGGGAUGCAGCCGCCUCGGCAGCUGCCAUCUGGGAAUCGCUGGCUGCCUUGGCUUCC .(((...((((((.....))))))...))).(((..((((((((((...((((...((-..((..(((((.((....)).)))))..))..))))))))))))))))..))).... ( -58.80, z-score = -2.66, R) >droEre2.scaffold_4770 9619525 115 + 17746568 UGCUGCCUCCGCCGCCCGAGCGGAUUGAGCUGCCUUGGCGGCCAGUUCGGAUUUGACC-UGGGAUGCAGCCGCCUCGGCUGCUGCCAUCUGGGAAUCGCUGGCUGCCUUCGCCUCC .(((...((((((....).)))))...))).((...((((((((((...((((...((-..((..(((((.((....)).)))))..))..))))))))))))))))...)).... ( -51.80, z-score = -1.06, R) >droVir3.scaffold_13047 18142405 115 + 19223366 CGCUGCCUCUGCGGCGCGAGCGGAUUGAGCCGCCCUCUCGGCCAGCUCCUGCUUGACC-UGCGUGGCAGCUGCCUCUGCGGCGGCCAUUUGUGCAUCACUGGCAGCCCUCGCCUCC .((((((...((((.((((((((((((.((((......))))))).))).))))).))-)))(((((.(((((....))))).)))))............)))))).......... ( -51.40, z-score = -0.58, R) >droMoj3.scaffold_6540 20561826 115 - 34148556 CGCUGCCUCGGCGGCACGUGCCGACUGCGCCGCCCGCUCGGCCAACUGCUGCUUGACC-UGUGUGGCAGCUGCCUCUGCGGCGGCCAUUUGCGCAUCGCUGGCGGCCCUCGCCUCC .(((((....))))).((((((((.((.((((((.((..(((...((((..(......-...)..))))..)))...)))))))))).))).))).))..((((.....))))... ( -53.00, z-score = -0.41, R) >droGri2.scaffold_15074 5747217 115 + 7742996 UGCCGCCUCCGCUGCUUUGGCAGCUGCAUAUGCUCGAUCCGCCAGCUGCUUCUUAACCAUGCG-GGCGACCUCCGCGGCAGCAGCCUGUUGUGUUCCAUAUGCCUGCUCUGCCUCG ...((((..(((((.((.((((((((.....((.......))))))))))....)).)).)))-)))).....((.(((((.(((..((((((...)))).))..)))))))).)) ( -37.00, z-score = 0.38, R) >anoGam1.chr2R 10468409 115 + 62725911 GGCGGCCUGGGCCGUGGCGGCCGCCUGGGAAGCCGCCUGGGCGAGCGUGUUCUGCGCCACGUA-CGCAGCCUGCUUGGCUGCGGCAUGCUGGUUAGCGACGGCCGAGUUGGCCUGA (((.(.((.((((((.(((((((((..((......))..))).(((((((..((((...))))-(((((((.....)))))))))))))))))).)).)))))).)).).)))... ( -67.10, z-score = -1.76, R) >apiMel3.GroupUn 75929369 115 + 399230636 UGCUGCAGCCGCAGUUGCCGCGGAUUGAGCAGCAGCUUGCGCCAAUGUGUUUUUCGCAACGAA-CGCAGCUUGUCCGGCCGCGGAAUGUUGAUUGCUCAAUUGCGUGUGCGCUUGU .(((((....))))).((((((((((((((((((((((.(((...(((((((........)))-))))(((.....))).))).)).)))).)))))))))).)))).))...... ( -43.30, z-score = 0.68, R) >consensus UGCUGCCUCCGCCGCCCGAGCGGAUUGAGCUGCCUUGGCGGCCAGUUCGUUUUUGACC_UGGGAUGCAGCCGCCUCGGCAGCUGCCAUCUGCGAAUCGCUGGCUGCCUUGGCCUCC .((.(((.((((.......)))).....(((((....(((((..........................)))))....)))))..................))).)).......... (-19.46 = -19.95 + 0.49)

| Location | 10,086,500 – 10,086,615 |
|---|---|
| Length | 115 |
| Sequences | 10 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 67.57 |
| Shannon entropy | 0.70679 |
| G+C content | 0.67217 |
| Mean single sequence MFE | -51.56 |
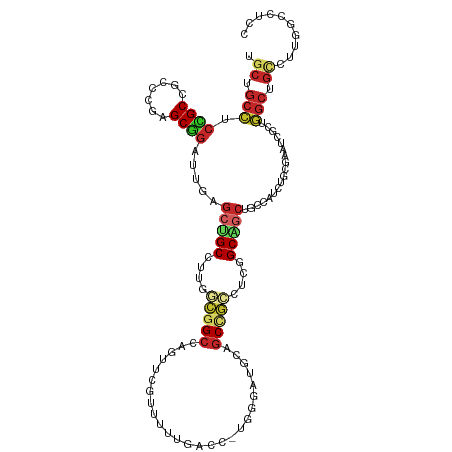
| Consensus MFE | -23.64 |
| Energy contribution | -23.43 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.69 |
| Mean z-score | -0.81 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.758813 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
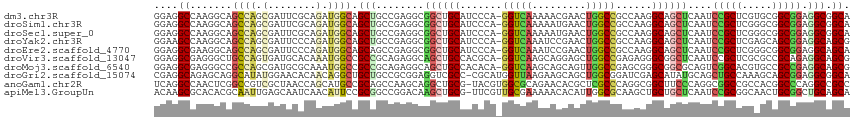
>dm3.chr3R 10086500 115 - 27905053 GGAGGCCAAGGCAGCCAGCGAUUCGCAGAUGGCAGCUGCCGAGGCGGCUGCAUCCCA-GGUCAAAAACGAACUGGCCGCCAAGGCAGCUCAAUCCGCUCGUGCGGCGGAGGCGGCA ....((....)).(((.((..(((((.(((.(.(((((((..((((((.......((-(.((......)).)))))))))..)))))))).)))(((....)))))))).))))). ( -51.81, z-score = -0.55, R) >droSim1.chr3R 16152480 115 + 27517382 GGAGGCCAAGGCAGCCAGCGAUUCGCAGAUGGCAGCUGCCGAGGCGGCUGCAUCCCA-GGUCAAAAAUGAACUGGCCGCCAAGGCAGCUCAAUCCGCUCGGGCGGCGGAGGCGGCA ....(((..((((((..((.(((....))).)).))))))..))).(((((.(((((-(.(((....))).)))((((((..(((..........)))..))))))))).))))). ( -56.10, z-score = -1.52, R) >droSec1.super_0 9166967 115 + 21120651 GGAGGCCAAGGCAGCCAGCGAUUCGCAGAUGGCAGCUGCCGAGGCGGCUGCAUCCCA-GGUCAAAAAUGAACUGGCCGCCAAGGCAGCUCAAUCCGCUCGGGCGGCGGAGGCGGCA ....(((..((((((..((.(((....))).)).))))))..))).(((((.(((((-(.(((....))).)))((((((..(((..........)))..))))))))).))))). ( -56.10, z-score = -1.52, R) >droYak2.chr3R 12470708 115 + 28832112 GGAAGCCAAGGCAGCCAGCGAUUCCCAGAUGGCAGCUGCCGAGGCGGCUGCAUCCCA-GGUCAAAUCCGAACUGGCCGCCAAGGCAGCUCAAUCCGCUCGAGCAGCGGAGGCAGCG ....(((..(((.(((((.........((((.(((((((....)))))))))))...-((......))...))))).)))..))).(((...((((((.....))))))...))). ( -53.30, z-score = -2.04, R) >droEre2.scaffold_4770 9619525 115 - 17746568 GGAGGCGAAGGCAGCCAGCGAUUCCCAGAUGGCAGCAGCCGAGGCGGCUGCAUCCCA-GGUCAAAUCCGAACUGGCCGCCAAGGCAGCUCAAUCCGCUCGGGCGGCGGAGGCAGCA ((..((....))..)).((..(((((((.(((..(((((((...)))))))...)))-((......))...)))((((((..(((..........)))..))))))))))...)). ( -51.10, z-score = -0.56, R) >droVir3.scaffold_13047 18142405 115 - 19223366 GGAGGCGAGGGCUGCCAGUGAUGCACAAAUGGCCGCCGCAGAGGCAGCUGCCACGCA-GGUCAAGCAGGAGCUGGCCGAGAGGGCGGCUCAAUCCGCUCGCGCCGCAGAGGCAGCG ....(((((((((((..(((...)))...((((((((.....))).((......)).-))))).))))((((((.((....)).))))))...)).)))))(((.....))).... ( -53.00, z-score = -0.23, R) >droMoj3.scaffold_6540 20561826 115 + 34148556 GGAGGCGAGGGCCGCCAGCGAUGCGCAAAUGGCCGCCGCAGAGGCAGCUGCCACACA-GGUCAAGCAGCAGUUGGCCGAGCGGGCGGCGCAGUCGGCACGUGCCGCCGAGGCAGCG ...(((....)))(((((((...)))...)))).(((.....))).((((((.....-((((((.......)))))).....((((((((.((....))))))))))..)))))). ( -52.90, z-score = 0.48, R) >droGri2.scaffold_15074 5747217 115 - 7742996 CGAGGCAGAGCAGGCAUAUGGAACACAACAGGCUGCUGCCGCGGAGGUCGCC-CGCAUGGUUAAGAAGCAGCUGGCGGAUCGAGCAUAUGCAGCUGCCAAAGCAGCGGAGGCGGCA ((.(((((....(((...((........)).))).))))).))...((((((-(((((((((.....((.....))......))).))))).(((((....))))).).)))))). ( -44.40, z-score = -0.34, R) >anoGam1.chr2R 10468409 115 - 62725911 UCAGGCCAACUCGGCCGUCGCUAACCAGCAUGCCGCAGCCAAGCAGGCUGCG-UACGUGGCGCAGAACACGCUCGCCCAGGCGGCUUCCCAGGCGGCCGCCACGGCCCAGGCCGCC ...((((.....((((((.(((....)))....(((((((.....)))))))-...(.((((.((......))))))).(((((((.(....).)))))))))))))..))))... ( -56.20, z-score = -1.73, R) >apiMel3.GroupUn 75929369 115 - 399230636 ACAAGCGCACACGCAAUUGAGCAAUCAACAUUCCGCGGCCGGACAAGCUGCG-UUCGUUGCGAAAAACACAUUGGCGCAAGCUGCUGCUCAAUCCGCGGCAACUGCGGCUGCAGCA ....((((...(((((((((....)))......((((((.......))))))-...)))))).....(.....))))).....(((((.....(((((....).))))..))))). ( -40.70, z-score = -0.10, R) >consensus GGAGGCCAAGGCAGCCAGCGAUUCGCAGAUGGCAGCUGCCGAGGCGGCUGCAUCCCA_GGUCAAAAACGAACUGGCCGACAAGGCAGCUCAAUCCGCUCGAGCGGCGGAGGCAGCA ....((.......((((.(........).)))).(((........((((((.......(((((.........)))))......))))))....(((((.....))))).))).)). (-23.64 = -23.43 + -0.21)
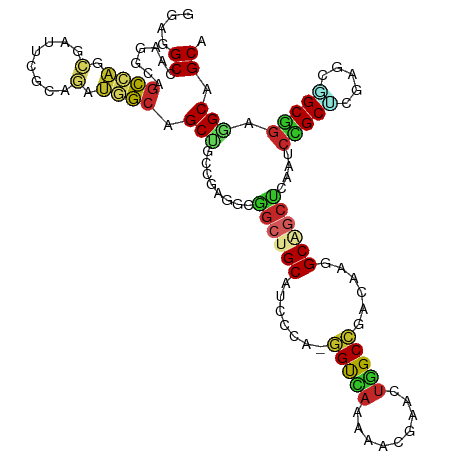
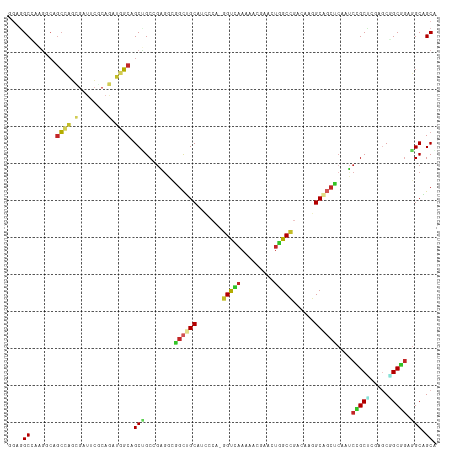
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:14:34 2011