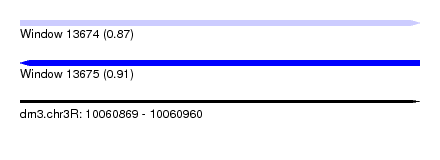
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 10,060,869 – 10,060,960 |
| Length | 91 |
| Max. P | 0.907555 |

| Location | 10,060,869 – 10,060,960 |
|---|---|
| Length | 91 |
| Sequences | 10 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 82.06 |
| Shannon entropy | 0.35533 |
| G+C content | 0.45354 |
| Mean single sequence MFE | -27.69 |
| Consensus MFE | -20.10 |
| Energy contribution | -21.30 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.871830 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
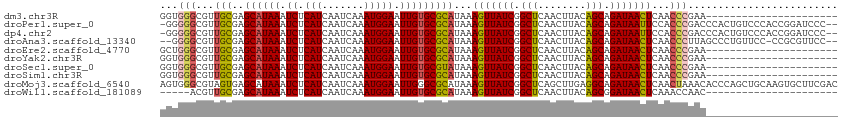
>dm3.chr3R 10060869 91 + 27905053 ----------------------UUCGGGUUGAGUUAUCUGCUGUAAGUUGAGCCGAUAACUUUAUGCGCACAAUUCCAUUUGAUUGAUGAGAUUUAUGCUCGCAACGCCCACC ----------------------...((((.((((((((.(((........))).))))))))..((((((.((.((((((.....)))).)).)).)))..)))..))))... ( -26.10, z-score = -2.53, R) >droPer1.super_0 7115325 110 + 11822988 --GGGAUCCGGUGGGACAGUGGGUCGGGUGGAAUUAUCUGCUGUAAGUUGAGCCGAUAACUUUAUGCGCACAAUUCCAUUUGAUUGAUGAGAUUUAUGCUCGCAACGCCCCC- --(((.....(((((.((((.((((((((((((((.(.(((...((((((......))))))...))).).)))))))))))))).))........)))))))....)))..- ( -32.40, z-score = -0.28, R) >dp4.chr2 17458402 110 - 30794189 --GGGAUCCGGUGGGACAGUGGGUCGGGUGGAAUUAUCUGCUGUAAGUUGAGCCGAUAACUUUAUGCGCACAAUUCCAUUUGAUUGAUGAGAUUUAUGCUCGCAACGCCCCC- --(((.....(((((.((((.((((((((((((((.(.(((...((((((......))))))...))).).)))))))))))))).))........)))))))....)))..- ( -32.40, z-score = -0.28, R) >droAna3.scaffold_13340 3290006 108 - 23697760 --GGAACGCGG-GGAACAGGGCUAAGGGUUGAGUUAUCUGCUGUAAGUUGAGCCGAUAACUUUAUGCGCACAAUUCCAUUUGAUUGAUGAGAUUUAUGCUCGCAACGCCCC-- --.......((-((....((((...(.((.((((((((.(((........))).))))))))...)).)..(((..((((.....))))..)))...))))......))))-- ( -30.00, z-score = -0.64, R) >droEre2.scaffold_4770 9596120 91 + 17746568 ----------------------UUCGGGUUGAGUUAUCUGCUGUAAGUUGAGCCGAUAACUUUAUGCGCACAAUUCCAUUUGAUUGAUGAGAUUUAUGCUCGCAACGCCCAGC ----------------------...((((.((((((((.(((........))).))))))))..((((((.((.((((((.....)))).)).)).)))..)))..))))... ( -26.10, z-score = -1.99, R) >droYak2.chr3R 12454009 91 - 28832112 ----------------------UUCGGGUUGAGUUAUCUGCUGUAAGUUGAGCCGAUAACUUUAUGCGCACAAUUCCAUUUGAUUGAUGAGAUUUAUGCUCGCAACGCCCACC ----------------------...((((.((((((((.(((........))).))))))))..((((((.((.((((((.....)))).)).)).)))..)))..))))... ( -26.10, z-score = -2.53, R) >droSec1.super_0 9150638 91 - 21120651 ----------------------UUCGGGUUGAGUUAUCUGCUGUAAGUUGAGCCGAUAACUUUAUACGCACAAUUCCAUUUGAUUGAUGAGAUUUAUGCUCGCAACGCCCACC ----------------------...((((.((((((((.(((........))).)))))))).....(((.((.((((((.....)))).)).)).))).......))))... ( -24.30, z-score = -2.41, R) >droSim1.chr3R 16136136 91 - 27517382 ----------------------UUCGGGUUGAGUUAUCUGCUGUAAGUUGAGCCGAUAACUUUAUGCGCACAAUUCCAUUUGAUUGAUGAGAUUUAUGCUCGCAACGCCCACC ----------------------...((((.((((((((.(((........))).))))))))..((((((.((.((((((.....)))).)).)).)))..)))..))))... ( -26.10, z-score = -2.53, R) >droMoj3.scaffold_6540 20544979 113 - 34148556 GUCGAAGCACUUGCAGCUGGGUGUUUAGUUGAGUUAUCUGCCUCAAGCUGAGCCGAUAACUUUAUGCGCCCAAUUCCAUUUGAUUGAUGAGAUUUAUGCUCACUACGCCCACU ((((((((....))...(((((((......((((((((.((.((.....)))).))))))))...)))))))......))))))...((((.......))))........... ( -29.60, z-score = -1.22, R) >droWil1.scaffold_181089 10928114 86 + 12369635 ----------------------GUUGGUUUGAGUUAUCCGCUGUAAGUUGAGCCGAUAACUUUAUGCGCACAAUUCCAUUUGAUUGAUGAGAUUUAUGCUCGCAACGU----- ----------------------((((((..((((((((.(((........))).))))))))...))(((.((.((((((.....)))).)).)).)))...))))..----- ( -23.80, z-score = -2.21, R) >consensus ______________________UUCGGGUUGAGUUAUCUGCUGUAAGUUGAGCCGAUAACUUUAUGCGCACAAUUCCAUUUGAUUGAUGAGAUUUAUGCUCGCAACGCCCACC .........................((((.((((((((.(((........))).))))))))..((((((.((.((((((.....)))).)).)).)))..)))..))))... (-20.10 = -21.30 + 1.20)

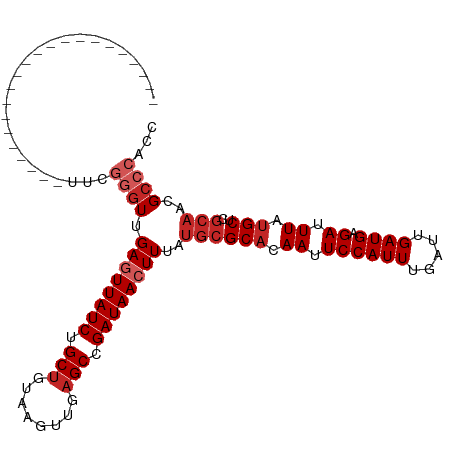
| Location | 10,060,869 – 10,060,960 |
|---|---|
| Length | 91 |
| Sequences | 10 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 82.06 |
| Shannon entropy | 0.35533 |
| G+C content | 0.45354 |
| Mean single sequence MFE | -25.06 |
| Consensus MFE | -20.52 |
| Energy contribution | -20.61 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.907555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 10060869 91 - 27905053 GGUGGGCGUUGCGAGCAUAAAUCUCAUCAAUCAAAUGGAAUUGUGCGCAUAAAGUUAUCGGCUCAACUUACAGCAGAUAACUCAACCCGAA---------------------- ..((((...(((..((((((.((.(((.......))))).)))))))))...(((((((.(((........))).)))))))...))))..---------------------- ( -23.80, z-score = -1.74, R) >droPer1.super_0 7115325 110 - 11822988 -GGGGGCGUUGCGAGCAUAAAUCUCAUCAAUCAAAUGGAAUUGUGCGCAUAAAGUUAUCGGCUCAACUUACAGCAGAUAAUUCCACCCGACCCACUGUCCCACCGGAUCCC-- -(((((.((((.(.((((((.((.(((.......))))).))))))......(((((((.(((........))).)))))))...).))))))...((((....)))))))-- ( -27.10, z-score = -0.52, R) >dp4.chr2 17458402 110 + 30794189 -GGGGGCGUUGCGAGCAUAAAUCUCAUCAAUCAAAUGGAAUUGUGCGCAUAAAGUUAUCGGCUCAACUUACAGCAGAUAAUUCCACCCGACCCACUGUCCCACCGGAUCCC-- -(((((.((((.(.((((((.((.(((.......))))).))))))......(((((((.(((........))).)))))))...).))))))...((((....)))))))-- ( -27.10, z-score = -0.52, R) >droAna3.scaffold_13340 3290006 108 + 23697760 --GGGGCGUUGCGAGCAUAAAUCUCAUCAAUCAAAUGGAAUUGUGCGCAUAAAGUUAUCGGCUCAACUUACAGCAGAUAACUCAACCCUUAGCCCUGUUCC-CCGCGUUCC-- --(((((..(((..((((((.((.(((.......))))).)))))))))...(((((((.(((........))).))))))).........))))).....-.........-- ( -28.60, z-score = -1.73, R) >droEre2.scaffold_4770 9596120 91 - 17746568 GCUGGGCGUUGCGAGCAUAAAUCUCAUCAAUCAAAUGGAAUUGUGCGCAUAAAGUUAUCGGCUCAACUUACAGCAGAUAACUCAACCCGAA---------------------- ....((.((((.(.((((((.((.(((.......))))).)))))).)....(((((((.(((........))).)))))))))))))...---------------------- ( -23.70, z-score = -1.78, R) >droYak2.chr3R 12454009 91 + 28832112 GGUGGGCGUUGCGAGCAUAAAUCUCAUCAAUCAAAUGGAAUUGUGCGCAUAAAGUUAUCGGCUCAACUUACAGCAGAUAACUCAACCCGAA---------------------- ..((((...(((..((((((.((.(((.......))))).)))))))))...(((((((.(((........))).)))))))...))))..---------------------- ( -23.80, z-score = -1.74, R) >droSec1.super_0 9150638 91 + 21120651 GGUGGGCGUUGCGAGCAUAAAUCUCAUCAAUCAAAUGGAAUUGUGCGUAUAAAGUUAUCGGCUCAACUUACAGCAGAUAACUCAACCCGAA---------------------- ....((.((((...((((((.((.(((.......))))).))))))......(((((((.(((........))).)))))))))))))...---------------------- ( -23.50, z-score = -1.81, R) >droSim1.chr3R 16136136 91 + 27517382 GGUGGGCGUUGCGAGCAUAAAUCUCAUCAAUCAAAUGGAAUUGUGCGCAUAAAGUUAUCGGCUCAACUUACAGCAGAUAACUCAACCCGAA---------------------- ..((((...(((..((((((.((.(((.......))))).)))))))))...(((((((.(((........))).)))))))...))))..---------------------- ( -23.80, z-score = -1.74, R) >droMoj3.scaffold_6540 20544979 113 + 34148556 AGUGGGCGUAGUGAGCAUAAAUCUCAUCAAUCAAAUGGAAUUGGGCGCAUAAAGUUAUCGGCUCAGCUUGAGGCAGAUAACUCAACUAAACACCCAGCUGCAAGUGCUUCGAC ..((((.((.(((.((...(((..(((.......)))..)))..)).)))..((((((((.(((.....))).).))))))).......)).))))(..((....))..)... ( -27.50, z-score = -0.09, R) >droWil1.scaffold_181089 10928114 86 - 12369635 -----ACGUUGCGAGCAUAAAUCUCAUCAAUCAAAUGGAAUUGUGCGCAUAAAGUUAUCGGCUCAACUUACAGCGGAUAACUCAAACCAAC---------------------- -----..((((.(.((((((.((.(((.......))))).))))))......(((((((.(((........))).)))))))....)))))---------------------- ( -21.70, z-score = -3.18, R) >consensus GGUGGGCGUUGCGAGCAUAAAUCUCAUCAAUCAAAUGGAAUUGUGCGCAUAAAGUUAUCGGCUCAACUUACAGCAGAUAACUCAACCCGAA______________________ ...(((...(((..((((((.((.(((.......))))).)))))))))...(((((((.(((........))).)))))))...)))......................... (-20.52 = -20.61 + 0.09)
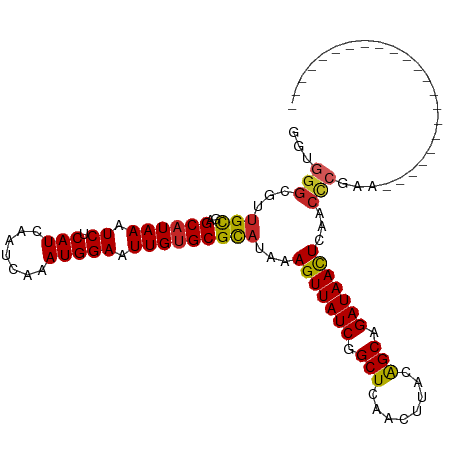
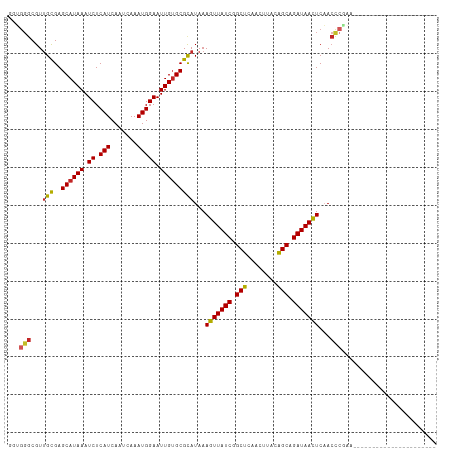
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:14:32 2011