
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 10,004,608 – 10,004,705 |
| Length | 97 |
| Max. P | 0.879578 |

| Location | 10,004,608 – 10,004,705 |
|---|---|
| Length | 97 |
| Sequences | 9 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 69.56 |
| Shannon entropy | 0.62525 |
| G+C content | 0.46275 |
| Mean single sequence MFE | -25.47 |
| Consensus MFE | -15.10 |
| Energy contribution | -14.78 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.54 |
| Mean z-score | -0.81 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.879578 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 10004608 97 - 27905053 ------------AGUGGAGUGGGCGCGAAACAGCUUAGAGAGCGACUCAAGUGGAAGAUUGGAUAUGAUAGUCAGACAUUGCGUAUACGCAAUGUGUUUCCUUUGGUUA ------------.((....(((((........)))))....))(((.((((.(((((((((.......)))))..((((((((....))))))))..))))))))))). ( -26.70, z-score = -1.26, R) >droPer1.super_0 7070697 97 - 11822988 ------------GUAGGAGUGGGAGUAGGGCCUCAUUGAGAGCAGCUGGAGUGGAAGAUUGGAUAUGAUAGUCGUACAUUGCGUAUACGCAGUGUGUUUCCCUUGGCUA ------------.....((((((.(.....)))))))......(((..(.(.(((((((((.......))))).(((((((((....))))))))).))))))..))). ( -28.30, z-score = -0.72, R) >dp4.chr2 17414196 97 + 30794189 ------------GUAGGAGUGGGAGUAGGGCCUCAUUGAGAGCAGCUGGAGUGGAAGAUUGGAUAUGAUAGUCGUACAUUGCGUAUACGCAGUGUGUUUCCCUUGGCUA ------------.....((((((.(.....)))))))......(((..(.(.(((((((((.......))))).(((((((((....))))))))).))))))..))). ( -28.30, z-score = -0.72, R) >droAna3.scaffold_13340 3241838 92 + 23697760 -----------------CGAGAAUAGGAAGCAGCUUCGAGAGCAACUCAAGUGGAAGAUUGGAUAUGAUAGUCAGACAUUGCGUAUACGCAGUGUGUUUCCUUUAGUUA -----------------.......(((((((.((((.(((.....)))))))....(((((.......)))))..((((((((....)))))))))))))))....... ( -26.00, z-score = -2.27, R) >droEre2.scaffold_4770 9553585 106 - 17746568 ---AGUGGAGUGGAGAAAGUGGGCGCGUAACAGCUUAGAGAGCGACUCAAGUGGAAGAUUGGAAAUGAUGGUCAGACGUUGCGUAUACGCAAUGUGUUUCCUUUGGGUA ---.((...(((...........)))...)).((((...)))).(((((((.(((((((((.......)))))..((((((((....))))))))..))))))))))). ( -27.20, z-score = -0.59, R) >droYak2.chr3R 12409445 109 + 28832112 UGUAGUGGAGCCAGUGGAGUGGGCGCGAAAGAGCUUAGAGAGCGACUCAAGUGGAAGAUUGGAUAUGAUAGUCAGACCUUGCGUAUACGCAAUGUGUUUCCUUUGGUUA ........((((((.(((((((((.(....).)))))....(((((.((((.(...(((((.......)))))...))))).))...))).......)))).)))))). ( -28.10, z-score = -0.18, R) >droSec1.super_0 9108719 97 + 21120651 ------------AGUGGAGUGGGCGCGAUACAGCUUAGAGAGCGACUCAAGUGGAAGAUUGGAUAUGAUAGUCAGACAUUGCGUAUACGCAAUGUGUUUCCUUUGGUUA ------------.(((..((....))..))).((((...))))(((.((((.(((((((((.......)))))..((((((((....))))))))..))))))))))). ( -27.20, z-score = -1.33, R) >droSim1.chr3R 16092508 97 + 27517382 ------------UGUGGAGUGGGCGCGAAACAGCUUAGAGAGCGACUCAAGUGGAAGAUUGGAUAUGAUAGUCAGACAUUGCGUAUACGCAAUGUGUUUCCUUUGGUUA ------------.((....(((((........)))))....))(((.((((.(((((((((.......)))))..((((((((....))))))))..))))))))))). ( -26.50, z-score = -1.15, R) >droGri2.scaffold_14846 100281 81 - 266023 -------------------------UGCGUCUAGUCAGUGAAUAAGCGUCUGCAAACGUGCCCCAAAACAGCC--ACAUUCAAUAUCAAGUAUACGCUUUAUUGGAAG- -------------------------.........(((((((...(((((.(((....(((.(........).)--))((.....))...))).))))))))))))...- ( -10.90, z-score = 0.98, R) >consensus ____________AGUGGAGUGGGCGCGAAACAGCUUAGAGAGCGACUCAAGUGGAAGAUUGGAUAUGAUAGUCAGACAUUGCGUAUACGCAAUGUGUUUCCUUUGGUUA ...........................................((((.(((.(((((((((.......)))))..((((((((....))))))))..))))))))))). (-15.10 = -14.78 + -0.33)

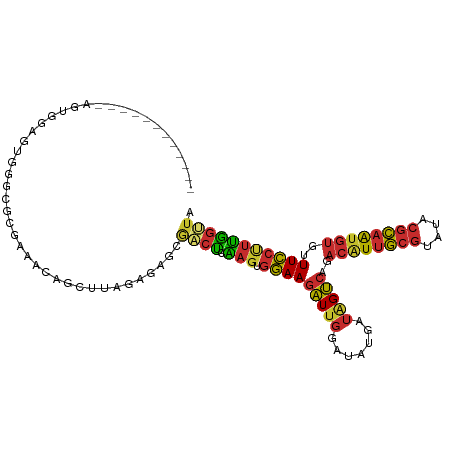
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:14:28 2011