| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,988,092 – 9,988,206 |
| Length | 114 |
| Max. P | 0.918393 |

| Location | 9,988,092 – 9,988,206 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 142 |
| Reading direction | forward |
| Mean pairwise identity | 62.61 |
| Shannon entropy | 0.60393 |
| G+C content | 0.49691 |
| Mean single sequence MFE | -31.55 |
| Consensus MFE | -16.59 |
| Energy contribution | -16.21 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.918393 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
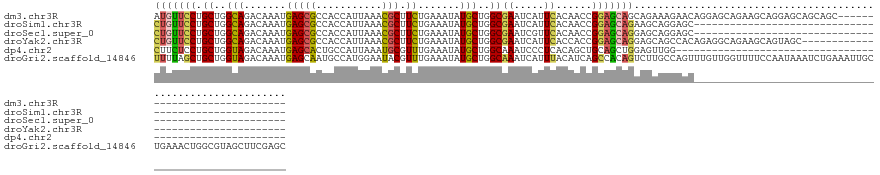
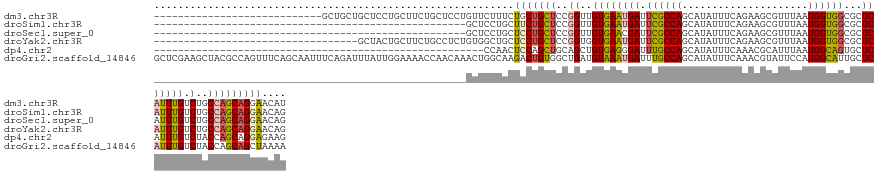
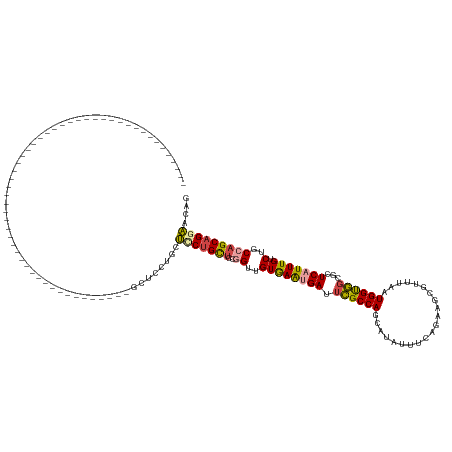
>dm3.chr3R 9988092 114 + 27905053 AUGUUCCUGCUGGCAGACAAAUGAGCGCCACCAUUAAACGCUUCUGAAAUAUGCUGGCGAAUCAUUCACAACCGGAGCAGCAGAAAGAACAGGAGCAGAAGCAGGAGCAGCAGC---------------------------- .(((((((((((((............)))..........(((((((.....(((((.(..................)))))).......)))))))...)))))))))).....---------------------------- ( -35.87, z-score = -1.80, R) >droSim1.chr3R 16075920 90 - 27517382 CUGUUCCUGCUGGCAGACAAAUGAGCGCCACCAUUAAACGCUUCUGAAAUAUGCUGGCGAAUCAUUCACAACCGGAGCAGAAGCAGGAGC---------------------------------------------------- ..(((((((((.((.......(((.(((((.(((......(....)....))).)))))..)))(((......)))))...)))))))))---------------------------------------------------- ( -27.20, z-score = -1.49, R) >droSec1.super_0 9092304 90 - 21120651 CUGUUCCUGCUGGCAGACAAAUGAGCGCCACCAUUAAACGCUUCUGAAAUAUGCUGGCGAAUCGUUCACAACCGGAGCAGGAGCAGGAGC---------------------------------------------------- (((((((((((((......(((((.(((((.(((......(....)....))).)))))..))))).....))..)))))))))))....---------------------------------------------------- ( -31.10, z-score = -2.09, R) >droYak2.chr3R 12391728 108 - 28832112 CUGUUCCUGCUGGCAGACAAAUGAGCGCCACCAUUAAACGCUUCUGAAAUAUGCUGGCGAAUCAUUCACCACCGGAGCAGGAGCAGCCACAGAGGCAGAAGCAGUAGC---------------------------------- ((((..(((.((((.....((((........))))....(((((((.....((.(((.((.....)).))).))...))))))).)))))))..))))..........---------------------------------- ( -33.90, z-score = -1.10, R) >dp4.chr2 17399095 87 - 30794189 CUUCUCCUGCUGGUAGACAAAUGAGCACUGCCAUUAAAUGCGUUUGAAAUAUGCUGGCAAAUCCCUCACAGCUGCAGCUGGAGUUGG------------------------------------------------------- ...((((.((((.(((.....((((...(((((......((((.......))))))))).....))))...))))))).))))....------------------------------------------------------- ( -24.30, z-score = -0.58, R) >droGri2.scaffold_14846 82089 142 + 266023 UUUUAGCUGCUGGUAGACAAAUGAGCAAUGCCAUGGAAUACGUUUGAAAUAUGCUGGCAAAUCAUUUACAUCAGCCACAGUCUUGCCAGUUUGUUGGUUUUCCAAUAAAUCUGAAAUUGCUGAAACUGGCGUAGCUUCGAGC ....((((((((((....((((((....(((((.....................)))))..))))))..))))((((((((..(..(((((((((((....)))))))).)))..)..))))....))))))))))...... ( -36.90, z-score = -0.46, R) >consensus CUGUUCCUGCUGGCAGACAAAUGAGCGCCACCAUUAAACGCUUCUGAAAUAUGCUGGCGAAUCAUUCACAACCGGAGCAGGAGCAGGAGC____________________________________________________ (((((((.((..(((.......(((((...........))).)).......)))..))((.....))......))))))).............................................................. (-16.59 = -16.21 + -0.38)
| Location | 9,988,092 – 9,988,206 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 142 |
| Reading direction | reverse |
| Mean pairwise identity | 62.61 |
| Shannon entropy | 0.60393 |
| G+C content | 0.49691 |
| Mean single sequence MFE | -31.87 |
| Consensus MFE | -17.92 |
| Energy contribution | -17.98 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.30 |
| Mean z-score | -0.95 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.850017 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
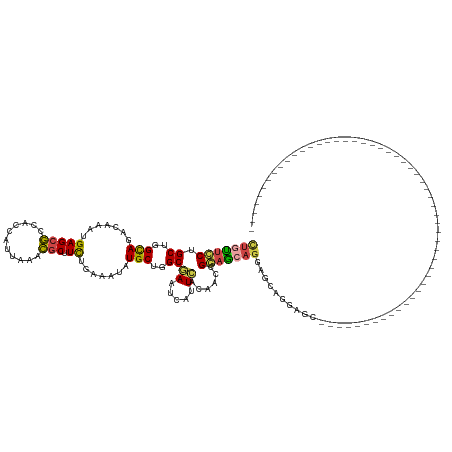
>dm3.chr3R 9988092 114 - 27905053 ----------------------------GCUGCUGCUCCUGCUUCUGCUCCUGUUCUUUCUGCUGCUCCGGUUGUGAAUGAUUCGCCAGCAUAUUUCAGAAGCGUUUAAUGGUGGCGCUCAUUUGUCUGCCAGCAGGAACAU ----------------------------((.(..((....))..).))...(((((...((((((...(((..(..(((((..(((((.(((....(......)....))).))))).)))))..)))).))))))))))). ( -32.80, z-score = 0.00, R) >droSim1.chr3R 16075920 90 + 27517382 ----------------------------------------------------GCUCCUGCUUCUGCUCCGGUUGUGAAUGAUUCGCCAGCAUAUUUCAGAAGCGUUUAAUGGUGGCGCUCAUUUGUCUGCCAGCAGGAACAG ----------------------------------------------------....(((.(((((((..(((.(..(((((..(((((.(((....(......)....))).))))).)))))..)..)))))))))).))) ( -30.80, z-score = -1.74, R) >droSec1.super_0 9092304 90 + 21120651 ----------------------------------------------------GCUCCUGCUCCUGCUCCGGUUGUGAACGAUUCGCCAGCAUAUUUCAGAAGCGUUUAAUGGUGGCGCUCAUUUGUCUGCCAGCAGGAACAG ----------------------------------------------------....(((.(((((((..(((.(..((.((..(((((.(((....(......)....))).))))).)).))..)..)))))))))).))) ( -29.40, z-score = -1.23, R) >droYak2.chr3R 12391728 108 + 28832112 ----------------------------------GCUACUGCUUCUGCCUCUGUGGCUGCUCCUGCUCCGGUGGUGAAUGAUUCGCCAGCAUAUUUCAGAAGCGUUUAAUGGUGGCGCUCAUUUGUCUGCCAGCAGGAACAG ----------------------------------(((((.((....))....)))))((.(((((((..(((((..(((((..(((((.(((....(......)....))).))))).)))))..)).)))))))))).)). ( -40.70, z-score = -1.97, R) >dp4.chr2 17399095 87 + 30794189 -------------------------------------------------------CCAACUCCAGCUGCAGCUGUGAGGGAUUUGCCAGCAUAUUUCAAACGCAUUUAAUGGCAGUGCUCAUUUGUCUACCAGCAGGAGAAG -------------------------------------------------------....((((.((((..((.(((((.(...(((((.....................))))).).)))))..))....)))).))))... ( -23.40, z-score = -0.22, R) >droGri2.scaffold_14846 82089 142 - 266023 GCUCGAAGCUACGCCAGUUUCAGCAAUUUCAGAUUUAUUGGAAAACCAACAAACUGGCAAGACUGUGGCUGAUGUAAAUGAUUUGCCAGCAUAUUUCAAACGUAUUCCAUGGCAUUGCUCAUUUGUCUACCAGCAGCUAAAA (((.((((((.....)))))))))....((((.(((.((((....)))).)))))))..((.((((((..(((..((((((..(((((..((((.......))))....)))))....)))))))))..))).))))).... ( -34.10, z-score = -0.55, R) >consensus ____________________________________________________GCUCCUGCUCCUGCUCCGGUUGUGAAUGAUUCGCCAGCAUAUUUCAGAAGCGUUUAAUGGUGGCGCUCAUUUGUCUGCCAGCAGGAACAG ............................................................(((((((..((..((((((((.((((((.....................))))))...))))))).)..))))))))).... (-17.92 = -17.98 + 0.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:14:25 2011