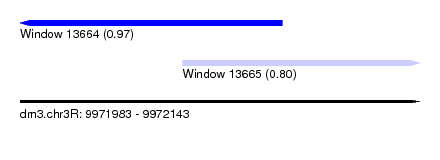
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,971,983 – 9,972,143 |
| Length | 160 |
| Max. P | 0.965777 |

| Location | 9,971,983 – 9,972,088 |
|---|---|
| Length | 105 |
| Sequences | 11 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 70.86 |
| Shannon entropy | 0.60114 |
| G+C content | 0.36722 |
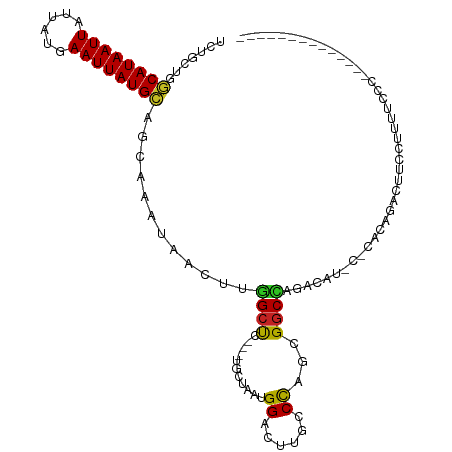
| Mean single sequence MFE | -18.61 |
| Consensus MFE | -9.93 |
| Energy contribution | -9.13 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.965777 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
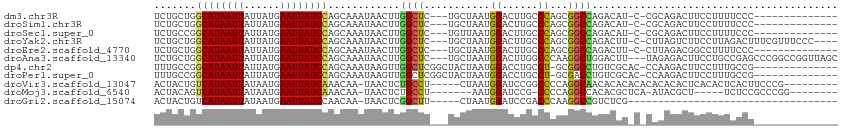
>dm3.chr3R 9971983 105 - 27905053 AGUUAUUUGCUGCAUAAUUCAUAAUAAUUAUGCCAGCAGAUUUGUGAUGCCCCC--AAAAACUUUU-C---AGCUGUUCAAAUUAAGCAGCGUCUUUUUGUGAAUACUCGU- ....(((((((((((((((......))))))).))))))))............(--(((((.....-.---.((((((.......))))))...))))))...........- ( -21.60, z-score = -1.45, R) >droSim1.chr3R 16059619 105 + 27517382 AGUUAUUUGCUGCAUAAUUCAUAAUAAUUAUGCCAGCAGAUUUGUGAUGCCCCC--AAAAACUUUU-C---AGCUGUUCAAAUUAAGCAGCGUCUUUUUGUGAAUACUCGU- ....(((((((((((((((......))))))).))))))))............(--(((((.....-.---.((((((.......))))))...))))))...........- ( -21.60, z-score = -1.45, R) >droSec1.super_0 9076159 105 + 21120651 AGUUAUUUGCUGCAUAAUUCAUAAUAAUUAUGCCGGCAGAUUUGUGAUGCCCCC--AAAAACUUUU-C---AGCUGUUCAAAUUAAGCAGCGUCUUUUUGUGAAUACUCGU- ...((((..(.((((((((......)))))))).((((.........))))...--..........-.---.((((((.......))))))........)..)))).....- ( -21.30, z-score = -0.96, R) >droYak2.chr3R 12375312 106 + 28832112 AGUUAUUUGCUGCAUAAUUCAUAAUAAUUAUGCCAGCAGAUUUGUGCUGCCCCC-AAAAAACUUUU-C---AGCUGUUCAAAUUAAGCAGCCUCUUUUUGUGAAUACUCGU- (((.(((((((((((((((......))))))).))))))))....)))......-...........-.---.((((((.......))))))....................- ( -21.80, z-score = -1.55, R) >droEre2.scaffold_4770 9520805 105 - 17746568 AGUUAUUUGCUGCAUAAUUCAUAAUAAUUAUGCCAGCAGAUUUGUGAUGCCCCC--AAAAACUUUU-C---AGCUGCUCAAAUUAAGCAGCGUCUUUUUGUGAAUACUCGU- ....(((((((((((((((......))))))).))))))))............(--(((((.....-.---.((((((.......))))))...))))))...........- ( -24.00, z-score = -2.04, R) >droAna3.scaffold_13340 3204597 106 + 23697760 AGUUAUUUGCUGCAUAAUUCAUAAUAAUUAUGCCAGCAGAUUUGUGAUGCCCCCCGAGAAACUUUC-C---AACUGUCCAAAUUAAGCUGCGUCGUUUUCUUAUAAAUAU-- ....(((((((((((((((......))))))).))))))))....(((((.....((((...))))-.---..((..........))..)))))................-- ( -17.70, z-score = -1.28, R) >dp4.chr2 17381676 99 + 30794189 ACUUAUUUGCUGCAUAAUUCAUAAUAAUUAUGCCGGCAAAUUUGUGCUGCCCCC---GAAACUUUCGC---AACUGUCCAAAUCAUUCUCCUUC-CUUACUUACCC------ .....((((((((((((((......))))))).)))))))((((..(......(---((.....))).---....)..))))............-...........------ ( -16.10, z-score = -3.22, R) >droPer1.super_0 7037827 99 - 11822988 ACUUAUUUGCUGCAUAAUUCAUAAUAAUUAUGCCGGCAAAUUUGUGCUGCCCCC---GAAACUUUCGC---AACUGUCCAAAUCAUUCUCCUUC-CUUACUUACCC------ .....((((((((((((((......))))))).)))))))((((..(......(---((.....))).---....)..))))............-...........------ ( -16.10, z-score = -3.22, R) >droVir3.scaffold_13047 18014766 99 - 19223366 AGUUA-UUGUUUCAUAAUUCAUUAUAAUUAUGACAGUAGUUUUGUAGAUGGCC---UAAAGCCUUGCC----GUCGUCCAAUGAUUUU-GAAGUUUUCCCCAAGUUGC---- (((((-(((..((((((((......)))))))).............((((((.---.........)))----)))...))))))))..-...................---- ( -17.00, z-score = -1.39, R) >droMoj3.scaffold_6540 20451904 103 + 34148556 AGUUA-UUGUUUCAUAAUUCAUUAUAAUUAUGACUGUAGUUGUGUAAAUGCCC---AGAGGCCGUGCC----GUUGCCCCAUCAUUUU-GAAGUUCCCUCGAAGUUGUGCCC .....-.....((((((((......))))))))..((((..(..(....(((.---...))).)..).----.))))..(((.(((((-((.......))))))).)))... ( -15.50, z-score = -0.19, R) >droGri2.scaffold_15074 1111521 108 + 7742996 AGUUA-UUGUUGCAUAAUUCAUUAUAAUUAUGACAGUAGUUUUACAAAUGCCC---AACAACCUUGCCACAAGCUGCCCAAUAAUUUUCCAAGUUUUUCACAAGUUAUUAAG (((((-(((.(((((((((......))))))).))(((((((..(((......---.......)))....))))))).)))))))).......................... ( -12.02, z-score = 0.02, R) >consensus AGUUAUUUGCUGCAUAAUUCAUAAUAAUUAUGCCAGCAGAUUUGUGAUGCCCCC__AAAAACUUUU_C___AGCUGUCCAAAUUAAGCAGCGUCUUUUUCUGAAUACUCGU_ ......(((((((((((((......)))))))).)))))......................................................................... ( -9.93 = -9.13 + -0.80)
| Location | 9,972,048 – 9,972,143 |
|---|---|
| Length | 95 |
| Sequences | 11 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 68.69 |
| Shannon entropy | 0.63625 |
| G+C content | 0.45979 |
| Mean single sequence MFE | -23.47 |
| Consensus MFE | -9.35 |
| Energy contribution | -9.00 |
| Covariance contribution | -0.35 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.801207 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9972048 95 + 27905053 UCUGCUGGCAUAAUUAUUAUGAAUUAUGCAGCAAAUAACUUGGCUC---UGCUAAUGGACUUGCCCAGCGGCCAGACAU-C-CGCAGACUUCCUUUUCCC-------------- ..(((((.(((((((......))))))))))))........((.((---(((..(((..((.(((....))).)).)))-.-.)))))...)).......-------------- ( -27.00, z-score = -2.13, R) >droSim1.chr3R 16059684 95 - 27517382 UCUGCUGGCAUAAUUAUUAUGAAUUAUGCAGCAAAUAACUUGGCUC---UGCUAAUGGACUUGCCCAGCGGCCAGACAU-C-CGCAGACUUCCUUUUCCC-------------- ..(((((.(((((((......))))))))))))........((.((---(((..(((..((.(((....))).)).)))-.-.)))))...)).......-------------- ( -27.00, z-score = -2.13, R) >droSec1.super_0 9076224 95 - 21120651 UCUGCCGGCAUAAUUAUUAUGAAUUAUGCAGCAAAUAACUUGGCUC---UGUUAAUGGACUUGCCCAGCGGCCAGACAU-C-CGCAGACUUCCUUUUCCC-------------- ..(((..((((((((......)))))))).)))........((.((---(((..(((..((.(((....))).)).)))-.-.)))))...)).......-------------- ( -23.40, z-score = -1.11, R) >droYak2.chr3R 12375378 105 - 28832112 UCUGCUGGCAUAAUUAUUAUGAAUUAUGCAGCAAAUAACUUGGCUC---UGCUAAUGGACUUGCCCAGCGGCCAGACUU-C-CUUAGUCUUCCUUAGACUUUCGUUUCCC---- ..(((((.(((((((......))))))))))))......((((((.---.(((...((......)))))))))))....-.-...(((((.....)))))..........---- ( -25.90, z-score = -1.48, R) >droEre2.scaffold_4770 9520870 95 + 17746568 UCUGCUGGCAUAAUUAUUAUGAAUUAUGCAGCAAAUAACUUGGCUC---UGCUAAUGGACUUGCCCAGCGGCCAGACUU-C-CUUAGACGGCCUUUUCCC-------------- ..(((((.(((((((......))))))))))))........((((.---..((((.(((((.(((....))).))...)-)-)))))..)))).......-------------- ( -28.00, z-score = -2.16, R) >droAna3.scaffold_13340 3204663 108 - 23697760 UCUGCUGGCAUAAUUAUUAUGAAUUAUGCAGCAAAUAACUUGGCUC---UGCUAAUGGACUUGGCCCAAGGCUGGACUU---UAGAGACUUCCUGCCGAGCCCGGCCGGUUAGC ..(((((.(((((((......))))))))))))..(((((.((((.---.(((..(((.......))).(((.(((...---........))).))).)))..))))))))).. ( -34.30, z-score = -1.16, R) >dp4.chr2 17381735 98 - 30794189 UUUGCCGGCAUAAUUAUUAUGAAUUAUGCAGCAAAUAAGUUGGCUCGGCUACUAAUGGACCUGCCU-GCGGCCUGUCGCAC-CCAAGACUUCCUUUGCCG-------------- .......((((((((......)))))))).(((((.((((((((..(((((....))).)).)))(-((((....))))).-....)))))..)))))..-------------- ( -26.80, z-score = -0.85, R) >droPer1.super_0 7037886 98 + 11822988 UUUGCCGGCAUAAUUAUUAUGAAUUAUGCAGCAAAUAAGUUGGCUCGGCUACUAAUGGACCUGCCU-GCGACCUGUCGCAC-CCAAGACUUCCUUUGCCG-------------- .......((((((((......)))))))).(((((.((((((((..(((((....))).)).)))(-((((....))))).-....)))))..)))))..-------------- ( -27.70, z-score = -2.15, R) >droVir3.scaffold_13047 18014826 99 + 19223366 ACUACUGUCAUAAUUAUAAUGAAUUAUGAAACAA-UAACUCUGCCU-----CUAAUGGAUCCGGCCCCAGGCAACACACACACACACACUCACACUCACUUCCCG--------- .......((((((((......)))))))).....-......(((((-----....((....)).....)))))................................--------- ( -12.30, z-score = -1.38, R) >droMoj3.scaffold_6540 20451968 91 - 34148556 ACUACAGUCAUAAUUAUAAUGAAUUAUGAAACAA-UAACUCUGCCU-------AAUGGAUCCG-CCCCAGGCCACACGCUCA-AUACGCU-----UCUCCGCCCGG-------- .......((((((((......)))))))).....-........((.-------...)).....-..((.(((.....((...-....)).-----.....))).))-------- ( -12.20, z-score = 0.05, R) >droGri2.scaffold_15074 1111590 73 - 7742996 ACUACUGUCAUAAUUAUAAUGAAUUAUGCAACAA-UAACUCGGCUU-----CUAAUGGAUCCGACCCAAGGCCGUCUCG----------------------------------- .....((.(((((((......)))))))))....-.....((((((-----....(((.......))))))))).....----------------------------------- ( -13.60, z-score = -1.66, R) >consensus UCUGCUGGCAUAAUUAUUAUGAAUUAUGCAGCAAAUAACUUGGCUC___UGCUAAUGGACUUGCCCAGCGGCCAGACAU_C_CACAGACUUCCUUUUCCC______________ .......((((((((......))))))))............((((........................))))......................................... ( -9.35 = -9.00 + -0.35)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:14:23 2011