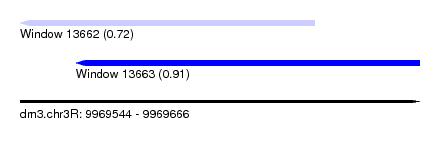
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,969,544 – 9,969,666 |
| Length | 122 |
| Max. P | 0.912490 |

| Location | 9,969,544 – 9,969,634 |
|---|---|
| Length | 90 |
| Sequences | 9 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 62.94 |
| Shannon entropy | 0.68994 |
| G+C content | 0.40679 |
| Mean single sequence MFE | -22.07 |
| Consensus MFE | -8.24 |
| Energy contribution | -8.57 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.716970 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3R 9969544 90 - 27905053 AUUUGGAUCCCCCUCCUCUGUGUGCUUGUGUUUAUUGGCAAAUAUUUUUGCUAGCUGAAAAAAUAUUUGCUCUCCAGUUCGAGCAGCAGU---------------- ....(((......))).((((.((((((........(((((((((((((.(.....).)))))))))))))........)))))))))).---------------- ( -25.09, z-score = -3.01, R) >droSim1.chr3R 16057540 88 + 27517382 -UUUGGA-ACCCCUCCUCUGUGUACUUGUGUUUAUUGGCAAAUAUUUUUGCUAGCUGGAAAAAUAUUUGCGCUCCAGUUCGAGCAGCAGU---------------- -...(((-.....))).((((................((((((((((((.(.....).))))))))))))((((......)))).)))).---------------- ( -24.00, z-score = -1.85, R) >droSec1.super_0 9073754 85 + 21120651 -UUUGGACCCCCCUCCUCUGUGUGCUUGUGUUUAUUGGCAAAUAUUUUUGCUAGCUGGUGUCG----GGCGCUCCAGUUCGAGCAGCAGU---------------- -...(((......))).((((.((((((.......(((((((....)))))))(((((.((..----...)).))))).)))))))))).---------------- ( -25.90, z-score = -0.92, R) >droYak2.chr3R 12372900 87 + 28832112 -UUUGAA--CCCCGUCUCUGUGUGCUUGUGUUUAUUGGCAAAUAUUUUUGCUAGCUGAAAAAAUAUUUGCGCUCCAGUUCGAGCAGCAGU---------------- -......--........((((.((((((.........((((((((((((.(.....).)))))))))))).........)))))))))).---------------- ( -21.77, z-score = -1.44, R) >droEre2.scaffold_4770 9518415 81 - 17746568 ---------GUGUGUCUCUGUGUGCUUGUGUUUAUUGGCAAAUAUUUUUGCUAGCUGAAAAAAUAUUUGCGCUCCAGUUCGAGCAGCAGU---------------- ---------........((((.((((((.........((((((((((((.(.....).)))))))))))).........)))))))))).---------------- ( -21.77, z-score = -1.44, R) >droAna3.scaffold_13340 3201488 102 + 23697760 UGUGCUCGAGUUUGUGUGUUAGCGUGUGUGUUUA-UUGCAAAUAUUUUUGCUUGCUUCAAAAAUAUUUGUCCUCCU---AGAGGAGCAAUGGCAGUCGAUUUCAGU ...(((.(((.(((..(((((...(((.......-...((((((((((((.......))))))))))))(((((..---.)))))))).)))))..))).)))))) ( -23.40, z-score = -0.59, R) >dp4.chr2 17379318 86 + 30794189 -----------UGCUUUCUCUAUGUGUGUGAUUG---GCAAAUAUUUUUGCUUGCUUGGAAAAUAUUUGUGCCACUAGAAGAGGAGCAAUGGCUGUGAAU------ -----------(((((((((((.(((.((.....---((((((((((((.(......))))))))))))))))))))).))).)))))............------ ( -20.30, z-score = -0.85, R) >droPer1.super_0 7035472 86 - 11822988 -----------UGCUUUCUCUACGUGUGUGAUUG---GCAAAUAUUUUUGCUUGCUUGGAAAAUAUUUGUGCCACUUGAAGAGGAGCAAUGGCUGUGAAU------ -----------(((((.((((.((.(((.(....---((((((((((((.(......))))))))))))).)))).)).)))))))))............------ ( -21.80, z-score = -1.43, R) >droWil1.scaffold_181089 10845985 72 - 12369635 ------------UGCGUGUGUGUGUUUAUG-CCAUUGGCAAAUAUUUUUGCUUGCUUAAAAAAUAUUUGCACUAUAG--AAAACAGU------------------- ------------.(((((........))))-).....((((((((((((.........)))))))))))).......--........------------------- ( -14.60, z-score = -0.22, R) >consensus __U______CCCCGUCUCUGUGUGCUUGUGUUUAUUGGCAAAUAUUUUUGCUAGCUGAAAAAAUAUUUGCGCUCCAGUUCGAGCAGCAGU________________ .....................................((((((((((((.........)))))))))))).................................... ( -8.24 = -8.57 + 0.32)
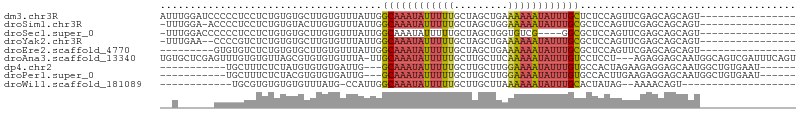
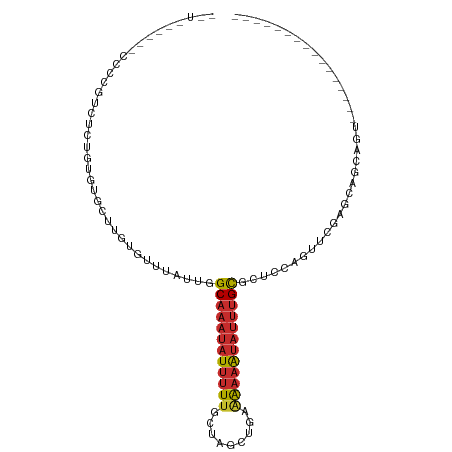
| Location | 9,969,561 – 9,969,666 |
|---|---|
| Length | 105 |
| Sequences | 8 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 65.98 |
| Shannon entropy | 0.71417 |
| G+C content | 0.42081 |
| Mean single sequence MFE | -21.15 |
| Consensus MFE | -6.93 |
| Energy contribution | -8.01 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.912490 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9969561 105 - 27905053 UAUACUUCGCCACUUUCGCCUCCGUCCCGGCCAUUUGGAUCCCCCUCCUCUGUGUGCUUGUGUUUAUUGGCAAAUAUUUUUGCUAGCUGAAAAAAUAUUUGCUCU .((((...(((((....(((........))).....(((......)))...))).))..)))).....(((((((((((((.(.....).))))))))))))).. ( -23.60, z-score = -2.84, R) >droSim1.chr3R 16057557 102 + 27517382 UAAACUUCGCCACUUUCACCUACG-CCCGGCU-UUUGGA-ACCCCUCCUCUGUGUACUUGUGUUUAUUGGCAAAUAUUUUUGCUAGCUGGAAAAAUAUUUGCGCU ........((((....(((.((((-(..((..-...((.-...)).))...)))))...))).....))((((((((((((.(.....).)))))))))))))). ( -20.90, z-score = -0.98, R) >droSec1.super_0 9073771 100 + 21120651 UAAACUUCGCCACUUUCACCUACGUCCCGGCU-UUUGGACCCCCCUCCUCUGUGUGCUUGUGUUUAUUGGCAAAUAUUUUUGCUAGCUGGUGU----CGGGCGCU .......((((.....((((..(((((.....-...))))...........................(((((((....))))))))..)))).----..)))).. ( -23.30, z-score = -0.03, R) >droYak2.chr3R 12372917 102 + 28832112 UAAACUUCGCCACUACCACCUCCGUCUCUGCU-UUUGAA--CCCCGUCUCUGUGUGCUUGUGUUUAUUGGCAAAUAUUUUUGCUAGCUGAAAAAAUAUUUGCGCU (((((...(((((....((.....((......-...)).--....))....))).))....)))))...((((((((((((.(.....).))))))))))))... ( -17.80, z-score = -1.49, R) >droEre2.scaffold_4770 9518432 96 - 17746568 UAAACUUCGCCACUUUCACCUCCGACCCCUCUGUGUG---------UCUCUGUGUGCUUGUGUUUAUUGGCAAAUAUUUUUGCUAGCUGAAAAAAUAUUUGCGCU (((((...(((((...(((..(.((....)).).)))---------.....))).))....)))))...((((((((((((.(.....).))))))))))))... ( -19.80, z-score = -2.53, R) >droAna3.scaffold_13340 3201518 104 + 23697760 UAAACUUUGCCACUUUCACCUCUUUGUCUGUGUGUGCUCGAGUUUGUGUGUUAGCGUGUGUGUUUAUU-GCAAAUAUUUUUGCUUGCUUCAAAAAUAUUUGUCCU (((((((.(((((....((......))....))).))..)))))))......................-(((((((((((((.......)))))))))))))... ( -18.50, z-score = -1.28, R) >droWil1.scaffold_181089 10845997 88 - 12369635 ----UCUUGUGGAUUCUGCGUGUAUGUGGG---UGUG---------CGUGUGUGUGUUUAUG-CCAUUGGCAAAUAUUUUUGCUUGCUUAAAAAAUAUUUGCACU ----....((((.....(((..((..((..---....---------))..))..))).....-))))..((((((((((((.........))))))))))))... ( -22.80, z-score = -1.72, R) >droVir3.scaffold_13047 18012200 80 - 19223366 ----------------CAACGCGGGCAUUGCU-UGUGG------CUUGUCCGUAUGUCAAUGUC--UUGACAAAUGUUUUUGCUUACUUCAAAAAUAUUUACUCU ----------------....(((((((..((.-....)------).)))))))..(((((....--)))))(((((((((((.......)))))))))))..... ( -22.50, z-score = -3.72, R) >consensus UAAACUUCGCCACUUUCACCUCCGUCCCGGCU_UUUGGA__CCCCUCCUCUGUGUGCUUGUGUUUAUUGGCAAAUAUUUUUGCUAGCUGAAAAAAUAUUUGCGCU .....................................................................((((((((((((.........))))))))))))... ( -6.93 = -8.01 + 1.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:14:21 2011