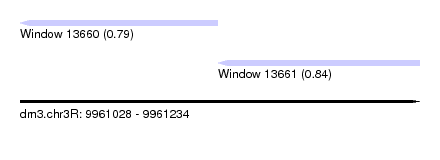
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,961,028 – 9,961,234 |
| Length | 206 |
| Max. P | 0.840167 |

| Location | 9,961,028 – 9,961,130 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 70.31 |
| Shannon entropy | 0.54293 |
| G+C content | 0.39260 |
| Mean single sequence MFE | -22.06 |
| Consensus MFE | -9.82 |
| Energy contribution | -9.94 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.791542 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
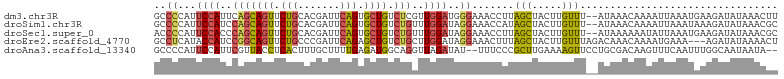
>dm3.chr3R 9961028 102 - 27905053 GCCCCAUUCCAUUCAGCAGUUCUGCACGAUUCAGUGCUGUCUCGUUGGAUGGGAAACCUUAGCUACUUGUUU--AUAAACAAAAUUAAAUGAAGAUAUAAACUU ......(((((((((((((....((((......))))...)).)))))))))))..............((((--(((..................))))))).. ( -23.57, z-score = -1.90, R) >droSim1.chr3R 16049147 102 + 27517382 GCCCCAUUCCAUCCAGCAGUUCUGCACGAUUCAGUGCUGUCUGUUUGGAUAGGAAACCAUAGCUACUUGUUU--AUAAACAAAAUUAAAUAAAGAUAUAAACGC ......(((((((((((((....((((......))))...))).)))))).)))).............((((--(((..................))))))).. ( -23.67, z-score = -2.61, R) >droSec1.super_0 9065362 102 + 21120651 ACCCCAUUCCACCCAGCAGUUCUGCACGAUUCAGUGCUGUCUGUUUGGAUAGGAAACCUUAGCUACUUGUUU--AUAAAAAAUAUUAAAUGAAGAUAUAAACGC ...((..((((..(((((((.(((.......))).)))).)))..))))..))...............((((--(((..................))))))).. ( -20.77, z-score = -1.55, R) >droEre2.scaffold_4770 9510142 101 - 17746568 GCCUCAUACCAUCCGGCAGUUCUGCCCGAUUCAGAGCUGUCUGCUUGGAUAGGAAACUUUAGCUACUUGUUUAGACAAACAAAAUGAAA---AGAUAUAAAACU ...((((.(((..(((((((((((.......)))))))).)))..)))..((....))........((((((....)))))).))))..---............ ( -23.10, z-score = -1.55, R) >droAna3.scaffold_13340 3192632 100 + 23697760 GCCCCAUUCCAUUCGUUACCUCACUUUGCUUUUGAGAUGGCAGGUUAGAUAU--UUUCCCGCUUGAAAAGUUCCUGCGACAAGUUUCAAUUUGGCAAUAAUA-- .........................(((((.(((((((.(((((.......(--((((......)))))...))))).....)))))))...))))).....-- ( -19.20, z-score = -0.47, R) >consensus GCCCCAUUCCAUCCAGCAGUUCUGCACGAUUCAGUGCUGUCUGGUUGGAUAGGAAACCUUAGCUACUUGUUU__AUAAACAAAAUUAAAUGAAGAUAUAAACGC ..((...((((..(((((((.(((.......))).)))).)))..))))..)).......(((.....)))................................. ( -9.82 = -9.94 + 0.12)

| Location | 9,961,130 – 9,961,234 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.69 |
| Shannon entropy | 0.33280 |
| G+C content | 0.39234 |
| Mean single sequence MFE | -25.80 |
| Consensus MFE | -17.52 |
| Energy contribution | -17.16 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.840167 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9961130 104 - 27905053 ----------AUAAAAGUGCUCCGCUCAA--GUGGCGCACGCAAAAUAACUUAAAAGCCGCAAAGUGGCUGGCAAAAACUAUUUAAGUUUAUGCGAUAACAACAAUAAAGGGACAG---- ----------......((((.((((....--)))).))))(((....((((((((((((((...))))))((......)).))))))))..)))......................---- ( -27.60, z-score = -2.24, R) >droSim1.chr3R 16049249 104 + 27517382 ----------AUAAAAGUGCUCCGCUCAA--GUGGCGCACGCAAAAUAACUUAAAAGCCGCAAAGUGGCUGGCAAAAACUAUUUAAGUUUAUGCGAUAACAACAAUAAAGGGACAG---- ----------......((((.((((....--)))).))))(((....((((((((((((((...))))))((......)).))))))))..)))......................---- ( -27.60, z-score = -2.24, R) >droSec1.super_0 9065464 104 + 21120651 ----------AUAAAAGUGCUCCGCUCAA--GUGGCGCACGCAAAAUAACUUAAAAGCCGCAAAGUGGCUGGCAAAAACUAUUUAAGUUUAUGCGAUAACAACAAUAAAGAGACAG---- ----------......((((.((((....--)))).)))).........(((...((((((...)))))).(((.(((((.....))))).)))...............)))....---- ( -27.80, z-score = -2.72, R) >droYak2.chr3R 12364528 104 + 28832112 ----------AUAAAAGUGCUCCGCUCAA--GUGGCGCACGCAAAAUAACUUAAAAGCCGCAAAGUGGCUGGCAAAAACUAUUUAAGUUUAUGCGAUAACAACAAUAAAGGGACAG---- ----------......((((.((((....--)))).))))(((....((((((((((((((...))))))((......)).))))))))..)))......................---- ( -27.60, z-score = -2.24, R) >droEre2.scaffold_4770 9510243 104 - 17746568 ----------AUAAAAGUGCUCCGCUCAA--GUGGCGCACGCAAAAUAACUUAAAAGCCGCAAAGUGGCUGGCAAAAACUAUUUAAGUUUAUGCGAUAACAACAAUAAAGGGACAG---- ----------......((((.((((....--)))).))))(((....((((((((((((((...))))))((......)).))))))))..)))......................---- ( -27.60, z-score = -2.24, R) >droAna3.scaffold_13340 3192732 114 + 23697760 GGGGCAUCAAAUAAAAGUGCUCCGCUCAA--GUGGUGCACGCAAAAUAACUUAAAAGCCGCAAAGUGGCUGGCAAAAACUAUUUAAGUUUAUGCGAUAACAACAAUAAAGGGACAG---- ..(..(((........((((.((((....--)))).))))(((....((((((((((((((...))))))((......)).))))))))..))))))..)................---- ( -30.40, z-score = -2.41, R) >dp4.chr2 17367601 113 + 30794189 -----CCAAAAUAAAAGUGCUUCACUCAA--GUGGUGCACGCAAAAUAACUUAAAAGCCGCAAAGUGGCUGGCAAAAACUAUUUAAGUUUAUGCGAUAACAACAGUUAAGAGGGCCCUUU -----((.........((((.((((....--)))).)))).........(((((.((((((...)))))).(((.(((((.....))))).)))...........))))).))....... ( -29.00, z-score = -1.80, R) >droPer1.super_0 7023661 113 - 11822988 -----CCAAAAUAAAAGUGCUUCACUCAA--GUGGUGCACGCAAAAUAACUUAAAAGCCGCAAAGUGGCUGGCAAAAACUAUUUAAGUUUAUGCGAUAACAACAGUUAAGAGGGCCCUUU -----((.........((((.((((....--)))).)))).........(((((.((((((...)))))).(((.(((((.....))))).)))...........))))).))....... ( -29.00, z-score = -1.80, R) >droWil1.scaffold_181089 10836148 91 - 12369635 -----------------CUCUUGACUUAA--GUGGUGCACGCAAAAUAACUUAAAAGCCACAAAGUGGCUGGCAAAAACUAUUUAAG-----GCGAUAAGAACAAUAAAAGCUCA----- -----------------.(((((.(((((--(((((....((.............((((((...)))))).))....))))))))))-----....)))))..............----- ( -20.94, z-score = -1.98, R) >droVir3.scaffold_13047 18001880 99 - 19223366 ----------------GUAGG-GACUUAAGAGUGCUGCACGCAAAAUAACUUAAAAGCCGCAAAGUGGCUGGUAAAAACUAUUUAAGUUUAUGCGAUAAGGACAAUAAAAGCACAG---- ----------------.....-.........(((((...((((....((((((((((((((...))))))(((....))).))))))))..))))..............)))))..---- ( -22.53, z-score = -1.41, R) >droMoj3.scaffold_6540 20438392 100 + 34148556 ----------------GUAGGCGACUUGAGAGUGCUGCACGCAAAAUAACUUAAAAACCGCAAAGUGGCUGGUAAAAACUAUUUAAGUUUAUGCGAUAAGGACAAUAAAAGCAGAG---- ----------------((((.(.(((....)))))))).((((....((((((((..((((...)))).((((....))))))))))))..)))).....................---- ( -18.20, z-score = 0.24, R) >droGri2.scaffold_15074 1098638 100 + 7742996 ----------------GUAAGCGACUUAAGAGUGCUGAACGCAAAAUAACUUAAAAGCCGCAAAGUGGCUGGCAAAAACUAUUUAAGUUUAUGCGAUAAGAACAAUAAAAGUACAG---- ----------------((((((.(((....))))))...((((....((((((((((((((...))))))((......)).))))))))..))))................)))..---- ( -21.30, z-score = -1.50, R) >consensus __________AUAAAAGUGCUCCACUCAA__GUGGUGCACGCAAAAUAACUUAAAAGCCGCAAAGUGGCUGGCAAAAACUAUUUAAGUUUAUGCGAUAACAACAAUAAAGGGACAG____ ................((((...((......)).)))).((((....((((((((((((((...))))))((......)).))))))))..))))......................... (-17.52 = -17.16 + -0.36)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:14:20 2011