
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,958,997 – 9,959,060 |
| Length | 63 |
| Max. P | 0.899231 |

| Location | 9,958,997 – 9,959,060 |
|---|---|
| Length | 63 |
| Sequences | 12 |
| Columns | 71 |
| Reading direction | reverse |
| Mean pairwise identity | 76.73 |
| Shannon entropy | 0.46248 |
| G+C content | 0.33442 |
| Mean single sequence MFE | -11.31 |
| Consensus MFE | -5.33 |
| Energy contribution | -5.44 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.899231 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3R 9958997 63 - 27905053 GUUACCUUACUUUUUUGGCCAUAAUUUCACAUUUAAAGCGAUUAAAUUUCGCUUUU-----GGCCUUG--- ................(((((.............(((((((.......))))))))-----))))...--- ( -14.41, z-score = -3.94, R) >droGri2.scaffold_15074 1096561 51 + 7742996 ------------GCUUGCAAAUAAUUUCACAUUUAAAGCGAUUAAAUUUCGC-UUGU----GGCCUUG--- ------------..............(((((.....(((((.......))))-))))----)).....--- ( -8.60, z-score = -0.65, R) >droMoj3.scaffold_6540 20436093 60 + 34148556 ---AUUUUUCUAGCUUGUAAAUAAUUUCACAUUUAAAGCGAUUAAAUUUCGC-UUGU----GGCCUUG--- ---.......................(((((.....(((((.......))))-))))----)).....--- ( -8.60, z-score = -0.89, R) >droVir3.scaffold_13047 17999194 57 - 19223366 -----AUUUUC-AUUUGGAAAUAAUUUCACAUUUAAAGCGAUUAAAUUUCGCUUGU-----GGCCUUG--- -----((((((-....))))))....(((((.....(((((.......))))))))-----)).....--- ( -10.70, z-score = -1.68, R) >droWil1.scaffold_181089 10834332 57 - 12369635 -----GUUGAC-UUUAUGAAAUAAUUUCACAUAUAAAGCGAUUAAAUUUCGCUAGU-----GCCCUUG--- -----....((-(...((((.....)))).......(((((.......))))))))-----.......--- ( -6.50, z-score = -0.61, R) >droPer1.super_0 7020800 66 - 11822988 -----GUUGCCAUUUAGGCCAUAAUUUCACAUUUAAAGCGAUUAAAUUUCGCUUGUUGACAACUCUUGUUA -----((((((.....)))................((((((.......))))))...)))........... ( -11.30, z-score = -1.40, R) >dp4.chr2 17365063 66 + 30794189 -----GCUGCCAUUUAGGCCAUAAUUUCACAUUUAAAGCGAUUAAAUUUCGCUUGUUGACAACUCUUGUUA -----...(((.....)))................((((((.......))))))..(((((.....))))) ( -11.30, z-score = -1.18, R) >droAna3.scaffold_13340 3190405 59 + 23697760 -----GUUUCUGGGUCCACCAUAAUUUCACAUUUAAAGCGAUUAAAUUUCGCUUUUU----GGCCUUG--- -----.....(((.....)))........((...(((((((.......))))))).)----)......--- ( -10.90, z-score = -1.73, R) >droYak2.chr3R 12362400 58 + 28832112 -----GUUACCUUUUCGGCCAUAAUUUCACAUUUAAAGCGAUUAAAUUUCGUUUUU-----GGCCUUG--- -----...........(((((.............(((((((.......))))))))-----))))...--- ( -12.31, z-score = -3.11, R) >droEre2.scaffold_4770 9507932 58 - 17746568 -----GUUACCUUUUCGGCCAUAAUUUCACAUUUAAAGCGAUUAAAUUUCGUUUUU-----GGCCUUG--- -----...........(((((.............(((((((.......))))))))-----))))...--- ( -12.31, z-score = -3.11, R) >droSec1.super_0 9063349 63 + 21120651 GUUACCAUUUUUUUUUGGCCAUAAUUUCACAUUUAAAGCGAUUAAAUUUCGCUUUU-----GGCCUUG--- ................(((((.............(((((((.......))))))))-----))))...--- ( -14.41, z-score = -3.75, R) >droSim1.chr3R 16047138 57 + 27517382 -----GUUACCUUUU-GGCCAUAAUUUCACAUUUAAAGCGAUUAAAUUUCGCUUUU-----GGCCUUG--- -----..........-(((((.............(((((((.......))))))))-----))))...--- ( -14.41, z-score = -3.70, R) >consensus _____GUUACCUUUUUGGCCAUAAUUUCACAUUUAAAGCGAUUAAAUUUCGCUUUU_____GGCCUUG___ ...................................((((((.......))))))................. ( -5.33 = -5.44 + 0.11)

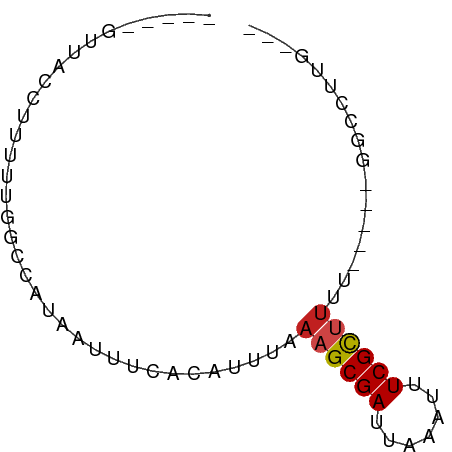
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:14:18 2011