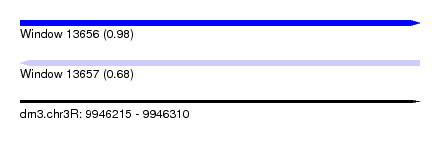
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,946,215 – 9,946,310 |
| Length | 95 |
| Max. P | 0.977502 |

| Location | 9,946,215 – 9,946,310 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 75.18 |
| Shannon entropy | 0.42214 |
| G+C content | 0.30522 |
| Mean single sequence MFE | -22.37 |
| Consensus MFE | -13.12 |
| Energy contribution | -14.32 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.977502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
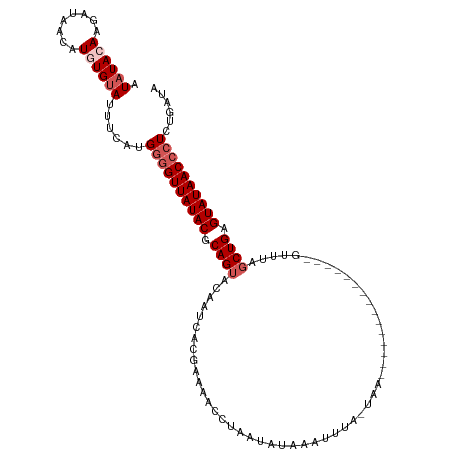
>dm3.chr3R 9946215 95 + 27905053 AUAUACAAGAUAACAUAUGAAUUUCAUGGAGUUAUACACAGUACAAACACGAAAACCUAGUAUAACUUUU--------------------UUUAGCUGAGUAUAACCCUAUGAUA .......................((((((.(((((((.((((............................--------------------....)))).))))))).)))))).. ( -16.75, z-score = -2.30, R) >droEre2.scaffold_4770 9495033 115 + 17746568 AUAUAGAAGAUAACAUGUGUAUUUCUUGGGGUUAUACGCAGUACAAUUACGAAAACCUAAUAUGAAUUUAAUAAAAAACUACUUUUCAUGUUUAUCUGAGUAUAACCCUCUGGUA ...(((((((..((....))...))))((((((((((.(((.................((((((((..................))))))))...))).)))))))))))))... ( -21.72, z-score = -1.26, R) >droYak2.chr3R 12349501 115 - 28832112 AUAUACAAUAUAAGAUGUGUAGUUCAUGGGGUUAUACGCAGUACGAUUACGAAAACCUAAUAUAAUUUUGAUAAAACACAACUUUUUGUGUUCAGCUGAGUAUAACCCUCUGAUA .((((((........))))))..(((.((((((((((.((((.......(((((...........)))))....(((((((....)))))))..)))).)))))))))).))).. ( -31.50, z-score = -3.84, R) >droSec1.super_0 9050532 92 - 21120651 AUAUACAAGAUAACAUGUGUAU--CAUGUGGUUAUACGCAGUACAAUCACGAAAACCUAGUAU-AACUUA-------------------GUUUAGCUGAGUAUAACCCUCAAUU- (((((((........)))))))--..((.((((((((.((((.(......).((((..((...-..))..-------------------)))).)))).))))))))..))...- ( -19.40, z-score = -1.80, R) >droSim1.chr3R 16034323 95 - 27517382 AUAUACAAGAUAACAUGUGUAUUUAAUGGGGUUAUACGCAGUACAAUCACGAAAACCUAUUAUAACUUUGUUAA--------------------GCUGAGUAUAACCCUCUGAUA (((((((........))))))).....((((((((((.((((((((.....................))))...--------------------)))).))))))))))...... ( -22.50, z-score = -2.46, R) >consensus AUAUACAAGAUAACAUGUGUAUUUCAUGGGGUUAUACGCAGUACAAUCACGAAAACCUAAUAUAAAUUUA_UAA_______________GUUUAGCUGAGUAUAACCCUCUGAUA .((((((........))))))......((((((((((.((((....................................................)))).))))))))))...... (-13.12 = -14.32 + 1.20)

| Location | 9,946,215 – 9,946,310 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 75.18 |
| Shannon entropy | 0.42214 |
| G+C content | 0.30522 |
| Mean single sequence MFE | -21.03 |
| Consensus MFE | -12.04 |
| Energy contribution | -12.08 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.681570 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
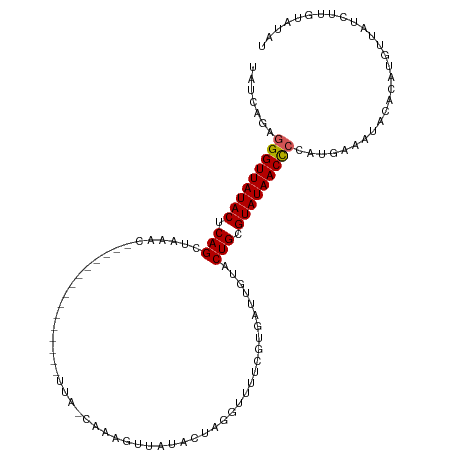
>dm3.chr3R 9946215 95 - 27905053 UAUCAUAGGGUUAUACUCAGCUAAA--------------------AAAAGUUAUACUAGGUUUUCGUGUUUGUACUGUGUAUAACUCCAUGAAAUUCAUAUGUUAUCUUGUAUAU ..((((.(((((((((.(((...((--------------------(...(..(........)..)...)))...))).))))))))).))))....................... ( -17.10, z-score = -1.03, R) >droEre2.scaffold_4770 9495033 115 - 17746568 UACCAGAGGGUUAUACUCAGAUAAACAUGAAAAGUAGUUUUUUAUUAAAUUCAUAUUAGGUUUUCGUAAUUGUACUGCGUAUAACCCCAAGAAAUACACAUGUUAUCUUCUAUAU .....(.(((((((((.(((((((..(((((((.((((((......))))).......).)))))))..)))).))).))))))))))((((...((....))..))))...... ( -22.91, z-score = -2.10, R) >droYak2.chr3R 12349501 115 + 28832112 UAUCAGAGGGUUAUACUCAGCUGAACACAAAAAGUUGUGUUUUAUCAAAAUUAUAUUAGGUUUUCGUAAUCGUACUGCGUAUAACCCCAUGAACUACACAUCUUAUAUUGUAUAU ..(((..(((((((((.(((..((((((((....))))))))(((.((((((......)))))).)))......))).)))))))))..))).....(((........))).... ( -28.20, z-score = -3.29, R) >droSec1.super_0 9050532 92 + 21120651 -AAUUGAGGGUUAUACUCAGCUAAAC-------------------UAAGUU-AUACUAGGUUUUCGUGAUUGUACUGCGUAUAACCACAUG--AUACACAUGUUAUCUUGUAUAU -....(((((((((((.(((..((((-------------------(.((..-...)).)))))...........))).))))))))(((((--.....)))))...)))...... ( -18.72, z-score = -0.56, R) >droSim1.chr3R 16034323 95 + 27517382 UAUCAGAGGGUUAUACUCAGC--------------------UUAACAAAGUUAUAAUAGGUUUUCGUGAUUGUACUGCGUAUAACCCCAUUAAAUACACAUGUUAUCUUGUAUAU .....(.(((((((((.(((.--------------------.(((((..(..((.....))..)..)).)))..))).)))))))))).....(((((..........))))).. ( -18.20, z-score = -0.88, R) >consensus UAUCAGAGGGUUAUACUCAGCUAAAC_______________UUA_CAAAGUUAUACUAGGUUUUCGUGAUUGUACUGCGUAUAACCCCAUGAAAUACACAUGUUAUCUUGUAUAU .......(((((((((.(((......................................................))).)))))))))............................ (-12.04 = -12.08 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:14:17 2011