| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,926,940 – 9,927,032 |
| Length | 92 |
| Max. P | 0.742666 |

| Location | 9,926,940 – 9,927,032 |
|---|---|
| Length | 92 |
| Sequences | 14 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 70.67 |
| Shannon entropy | 0.65581 |
| G+C content | 0.59576 |
| Mean single sequence MFE | -32.29 |
| Consensus MFE | -14.19 |
| Energy contribution | -14.35 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.742666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
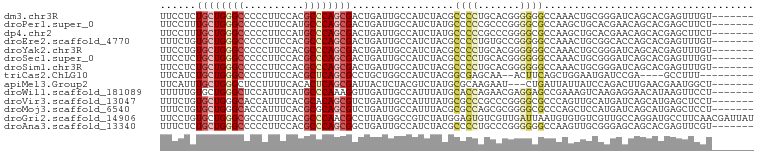
>dm3.chr3R 9926940 92 - 27905053 UUCCUCUGCUGGGCCCCCUUCCACGCCCAGCGACUGAUUGCCAUCUACGCCCCUGCACGGGGGGCCAAACUGCGGGAUCAGCACGAGUUUGU------- ...(((.(((((((..........)))))))(.((((((.(((.....((((((.....)))))).....)).).)))))))..))).....------- ( -36.10, z-score = -1.28, R) >droPer1.super_0 6989871 92 - 11822988 UUCCUUUGCUGGGCCCCCUUCCAUGCCCAGCGACUGAUUGCCAUCUAUGCCCCCGCCCGGGGCGCCAAGCUGCACGAACAGCACGAGCUUCU------- .((..(((((((((..........)))))))))..))..(((......(((((.....))))).....((((......))))..).))....------- ( -32.50, z-score = -1.75, R) >dp4.chr2 17334594 92 + 30794189 UUCCUUUGCUGGGCCCCCUUCCAUGCCCAGCGACUGAUUGCCAUCUAUGCCCCCGCCCGGGGCGCCAAGCUGCACGAACAGCACGAGCUUCU------- .((..(((((((((..........)))))))))..))..(((......(((((.....))))).....((((......))))..).))....------- ( -32.50, z-score = -1.75, R) >droEre2.scaffold_4770 9472408 92 - 17746568 UUUCUGUGCUGGGCCCCCUUCCACGCCCAGCGACUGAUUGCCAUCUACGCCCCUGUGCCGGGGGCCAAACUGCGGCACCAGCACGAGUUUGU------- ..((..((((((((..........))))))))...))...........(((((((...)))))))((((((.((((....)).)))))))).------- ( -36.00, z-score = -1.37, R) >droYak2.chr3R 12328402 92 + 28832112 UUCCUGUGCUGGGCCCCCUUCCACGCCCAGCGACUGAUUGCCAUCUACGCCCCUGCACGGGGGGCCAAACUGCGGGAUCAGCACGAGUUUGU------- .......(((((((..........)))))))(.((((((.(((.....((((((.....)))))).....)).).)))))))..........------- ( -33.80, z-score = -0.25, R) >droSec1.super_0 9031336 92 + 21120651 UUCCUCUGCUGGGCCCCCUUCCACGCCCAGCGACUGAUUGCCAUCUACGCCCCUGCACGGGGGGCCAAACUGCGGGAUCAGCACGAGUUUGU------- ...(((.(((((((..........)))))))(.((((((.(((.....((((((.....)))))).....)).).)))))))..))).....------- ( -36.10, z-score = -1.28, R) >droSim1.chr3R 16015268 92 + 27517382 UUCCUCUGCUGGGCCCCCUUCCACGCCCAGCGACUGAUUGCCAUCUACGCCCCUGCACGGGGGGCCAAACUGCGGGAUCAGCACGAGUUUGU------- ...(((.(((((((..........)))))))(.((((((.(((.....((((((.....)))))).....)).).)))))))..))).....------- ( -36.10, z-score = -1.28, R) >triCas2.ChLG10 4512953 84 + 8806720 UUCAUCUGCUGGGCCCCUUUCCACGCUCAGCGCCUGCUGGCCAUCUACGGCGAGCAA--ACUUCAGCUGGAAUGAUCCGA----GCCUUU--------- .......(((((((..........)))))))...((((.(((......))).)))).--......((((((....))).)----))....--------- ( -27.70, z-score = -0.98, R) >apiMel3.Group2 12020902 89 + 14465785 UUCAUUUGCUGGGCUCCUUUUCACACUCAGCGAUUACUCUACGUCUAUGCGCAAGAAU---CUGAUUAUUAUCCAGACUUGAACGAAUGGCU------- .....((((((((............)))))))).........(((..((..((((..(---(((.........))))))))..))...))).------- ( -16.80, z-score = -0.49, R) >droWil1.scaffold_181089 10804614 92 - 12369635 UUUUUGUGCUGGGCUCCAUUUCAUGCCCAAAGGUUGAUUGCCAUUUAUGCACCAGAACGAGGAGCCGAAAGUCAAGAGGAACAUAAGUUCCU------- .(((((.(((.((((((..(((.(((..((((((.....))).)))..)))...)))...))))))...))))))))(((((....))))).------- ( -27.40, z-score = -1.80, R) >droVir3.scaffold_13047 17964890 92 - 19223366 UUUCUGUGCUGGGCACCAUUUCACGCACAGCGUCUGAUUGCCAUUUAUGCGCCCGCCCGGGGCGCCCAGUUGCAUGAUCAGCAUGAGCUCCU------- ...((((((((((......)))).))))))(((((((((((.(((...((((((.....))))))..))).)))..))))).))).......------- ( -31.40, z-score = -0.33, R) >droMoj3.scaffold_6540 20403412 92 + 34148556 UUUCUGUGCUGGGCACCAUUUCACGCGCAGCGUCUGAUUGCCAUUUACGCGCCAGCGCGGGGCGCCCAGCUCCAUGAUCAGCAUGAGCUCCU------- .......(((((((.((......(((((.((((.(((.......))).))))..))))).)).)))))))..((((.....)))).......------- ( -36.00, z-score = -1.17, R) >droGri2.scaffold_14906 10480037 99 + 14172833 UUCCUGUGCUGGGCGCCAUUUCACGCCCAACGCCUUAUGGCCGUCUAUGGAGUGUCGUUGAUUAAUGUGUGUCGUUGCCAGGAUGCCUUCAACGAUUAU ((((((.((((((((........))))))..(((....))).)).)).)))).((((((((.....(.(((((........)))))).))))))))... ( -29.80, z-score = -0.83, R) >droAna3.scaffold_13340 3153298 92 + 23697760 UUUCUCUGCUGGGCCCCCUUCCACGCCCAGCGGCUGAUUGCCAUCUACGCCCCUGCCCGGGGGGCCAAGUUGCGGGAGCAGCACGAGUUCGU------- ...(((.(((((((..........)))))))(((.....)))......((((((.....))))))...(((((....)))))..))).....------- ( -39.90, z-score = -0.67, R) >consensus UUCCUGUGCUGGGCCCCCUUCCACGCCCAGCGACUGAUUGCCAUCUACGCCCCUGCACGGGGGGCCAAGCUGCGGGAUCAGCACGAGUUUCU_______ ......((((((((..........)))))))).................((((.......))))................................... (-14.19 = -14.35 + 0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:14:12 2011