| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,924,398 – 9,924,473 |
| Length | 75 |
| Max. P | 0.872790 |

| Location | 9,924,398 – 9,924,473 |
|---|---|
| Length | 75 |
| Sequences | 11 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 66.03 |
| Shannon entropy | 0.65513 |
| G+C content | 0.52883 |
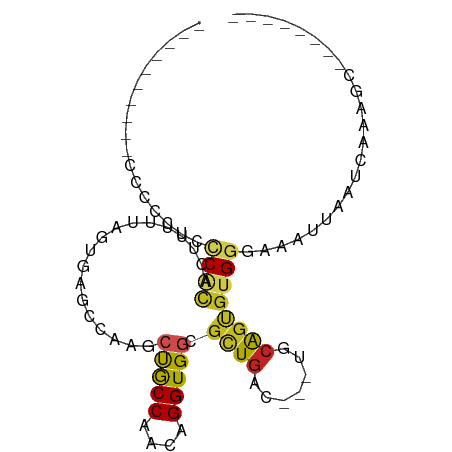
| Mean single sequence MFE | -27.06 |
| Consensus MFE | -7.75 |
| Energy contribution | -7.64 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.62 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.872790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
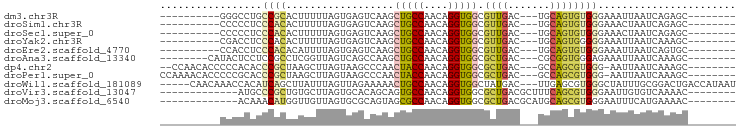
>dm3.chr3R 9924398 75 - 27905053 ----------GGGCCUGCCGCACUUUUUAGUGAGUCAAGCUGCCAACAGGUGGCGUUGAC---UGCAGUGUGGGAAAUUAAUCAGAGC-------- ----------..((((((((((((.....((.((((((((..((....))..)).)))))---)))))))))).........))).))-------- ( -26.10, z-score = -1.21, R) >droSim1.chr3R 16012597 75 + 27517382 ----------CCCCCUCCCACACUUUUUAGUGAGUCAAGCUGCCAACAGGUGGCGUUGAC---UGCAGUGUGGGAAACUAAUCAGAGC-------- ----------.....(((((((((.....((.((((((((..((....))..)).)))))---)))))))))))).............-------- ( -30.30, z-score = -3.39, R) >droSec1.super_0 9028700 75 + 21120651 ----------CCCCCUCCCACACUUUUUAGUGAGUCAAGCUGCCAACAGGUGGCGUUGAC---UGCAGUGUGGGAAACUAAUCAGAGC-------- ----------.....(((((((((.....((.((((((((..((....))..)).)))))---)))))))))))).............-------- ( -30.30, z-score = -3.39, R) >droYak2.chr3R 12325918 75 + 28832112 ----------CGACCUCCCACACUUUUUAGUGAGUCAAGCUGCCAACAGGUGGCGUUGAC---UGCAGUGGGGGAAAUUAAUCAAAGC-------- ----------....((((((((((....))).((((((((..((....))..)).)))))---)...)))))))..............-------- ( -28.80, z-score = -3.16, R) >droEre2.scaffold_4770 9469811 75 - 17746568 ----------CCACCUCCCACACAUUUUAGUGAGUCAAGCUGCCAACAGGUGGCGUUGAC---UGCAGUGUGGGAAAUUAAUCAGUGC-------- ----------.....((((((((.......((((((((((..((....))..)).)))))---).)))))))))).............-------- ( -28.61, z-score = -2.97, R) >droAna3.scaffold_13340 3150968 77 + 23697760 --------CAUACUCCUCCGCCUCGGUUAGUCAGCCAAGCUGCCAACAGGUGGCGCUGAC---CGCGGUGGGAGAAAUUAAUCAAAGC-------- --------.....((..(((((.((((((((..((((..(((....))).))))))))))---)).)))))..)).............-------- ( -33.20, z-score = -3.75, R) >dp4.chr2 17331591 82 + 30794189 --CCAACACCCCCACACCCGCUAAGCUUAGUAAGCCCAACUACCAACAGGUGGCGCUGAC---GCCAGCGUGGG-AAUUAAUCAAAGC-------- --........(((((....(((..(((.....)))......(((....))))))((((..---..)))))))))-.............-------- ( -21.50, z-score = -1.25, R) >droPer1.super_0 6986854 84 - 11822988 CCAAAACACCCCCGCACCCGCUAAGCUUAGUAAGCCCAACUACCAACAGGUGGCGCUGAC---GCCAGCGUGGG-AAUUAAUCAAAGC-------- ...................(((..(.(((((...((((.(((((....)))))(((((..---..)))))))))-.))))).)..)))-------- ( -21.30, z-score = -0.71, R) >droWil1.scaffold_181089 10801536 88 - 12369635 -----CAACAAACCACAUCAGCUUAUUUAGUUAGAAAAACUGCCAACAGGUGGCUAUGAC---UUGAGCGUGGGCUAUUUGCGGACUGACCAUAAU -----.......((((....(((((...(((((......(..((....))..)...))))---))))))))))((.....))((.....))..... ( -18.40, z-score = -0.27, R) >droVir3.scaffold_13047 17961372 75 - 19223366 -------------AUGCCCGCUGUGCUUAGUGCACAGCAGUGCCAACAGGUGGCGCUGACGCUUUCAGCGUGGGAAUUGUGUCAAAAC-------- -------------...((((((((((.....)))))))(((((((.....))))))).((((.....)))))))..............-------- ( -33.00, z-score = -2.48, R) >droMoj3.scaffold_6540 20400037 75 + 34148556 -------------ACAAACAUGGUUGUUAGUGCGCAGUAGCGCCAACAGGUGGCGCUGACGCAUGCAGCGUGGGAAUUUCAUGAAAAC-------- -------------.....((((.((((..(((((...((((((((.....)))))))).)))))))))))))................-------- ( -26.20, z-score = -1.54, R) >consensus __________CCCCCUCCCACACUUUUUAGUGAGCCAAGCUGCCAACAGGUGGCGCUGAC___UGCAGUGUGGGAAAUUAAUCAAAGC________ .................((((..................(((((....))))).((((.......))))))))....................... ( -7.75 = -7.64 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:14:11 2011