| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,911,122 – 9,911,217 |
| Length | 95 |
| Max. P | 0.606463 |

| Location | 9,911,122 – 9,911,217 |
|---|---|
| Length | 95 |
| Sequences | 9 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 82.26 |
| Shannon entropy | 0.37065 |
| G+C content | 0.42363 |
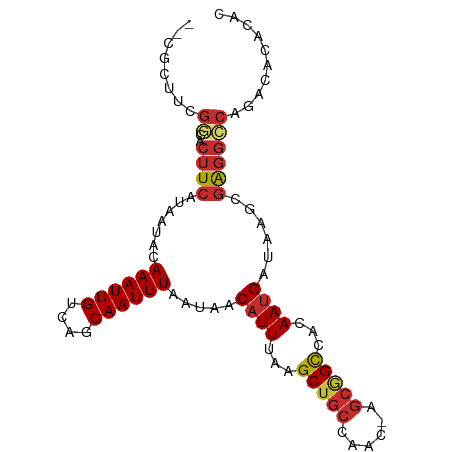
| Mean single sequence MFE | -18.80 |
| Consensus MFE | -12.32 |
| Energy contribution | -12.78 |
| Covariance contribution | 0.46 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.606463 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
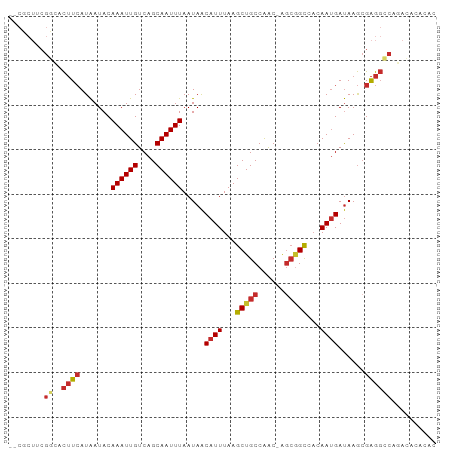
>dm3.chr3R 9911122 95 + 27905053 CGCACUUCGGCACUUCAUAAUACAAAUUGUCAGCAAUUUAAUAACAUUUAAGCUGCCAAC-AGCGGCCACAAUGAUAAGCGAGGCCAGACAAUCAC .((......))..............((((((....................((((....)-)))((((..............)))).))))))... ( -18.94, z-score = -1.51, R) >droSim1.chr3R 15998899 95 - 27517382 CGCUCUUCGGCACUUCAUAAUACAAAUUGUCAGCAAUUUAAUAACAUUUAAGCUGCCAAC-AGCGGCCACAAUGAUAAGCGAGGCCAGACAUACAC .(((.((.(((((((........((((((....))))))..........))).)))))).-)))((((..............)))).......... ( -18.81, z-score = -1.30, R) >droSec1.super_0 9015520 88 - 21120651 -------CGGCACUUCAUAAUACAAAUUGUCAGCAAUUUAAUAACAUUUAAGCUGCCAAC-AGCGGCCACAAUGAUAAGCGAGGCCAGACACACAC -------.(((.(((........((((((....)))))).....((((...(((((....-.)))))...))))......)))))).......... ( -18.60, z-score = -2.35, R) >droYak2.chr3R 12311675 95 - 28832112 CGCUCUUCGGCACUUCAUAAUACAAAUUGUCAGCAAUUUAAUAACAUUUAAGCUGCCAAC-AGCGGCCACAAUGAUAAGCGAGGCCAGACAUACAC .(((.((.(((((((........((((((....))))))..........))).)))))).-)))((((..............)))).......... ( -18.81, z-score = -1.30, R) >droEre2.scaffold_4770 9451898 95 + 17746568 CGCUCUUCGGCACUUCAUAAUACAAAUUGUCAGCAAUUUAAUAACAUUUAAGCUGCCAAC-AGCUGCCACAAUGAUAAGCGAGGCCAGACAUACAC ((((..(((((............((((((....))))))...........(((((....)-))))))).....))..))))............... ( -18.70, z-score = -1.31, R) >droAna3.scaffold_13340 3133019 87 - 23697760 --CGCUGCGCGACUUCAUAAUACAAAUUGUCAGCAAUUUAAUAACAUUUAAGCUGCCAAC-GUCGGUAACAAUGAUAAGCGAGGCCAGAC------ --.(((.(((....((((..(((....(((..(((.(((((......))))).)))..))-)...)))...))))...))).))).....------ ( -18.80, z-score = -0.94, R) >dp4.chr2 17317676 93 - 30794189 --CGCUUCGCGACUUCAUAAUACAAAUUGUCAGCAAUUUAAUAACAUUUAAGCUGCCAGCCAGCGGUAACAAUGAUAAGCGAGGGCACAGACGCA- --..((((((....((((..(((((((((....))))))............((((.....)))).)))...))))...))))))((......)).- ( -22.10, z-score = -1.26, R) >droPer1.super_0 6972785 93 + 11822988 --CGCUUCGCGACUUCAUAAUACAAAUUGUCAGCAAUUUAAUAACAUUUAAGCUGCCAGCCAGCGGUAACAAUGAUAAGCGAGGGCACAGACGCA- --..((((((....((((..(((((((((....))))))............((((.....)))).)))...))))...))))))((......)).- ( -22.10, z-score = -1.26, R) >droGri2.scaffold_15074 3279460 81 + 7742996 --------AAUUUAUAAUGAUACAAAUUGUCAGCAAUUUAAUAGCAUUUAAGCUGCCGACAGGAAGCU-CAACGACAAAAGGACACACAC------ --------..................(((((.((.........)).....((((.((....)).))))-....)))))............------ ( -12.30, z-score = -1.56, R) >consensus __CGCUUCGGCACUUCAUAAUACAAAUUGUCAGCAAUUUAAUAACAUUUAAGCUGCCAAC_AGCGGCCACAAUGAUAAGCGAGGCCAGACACACAC ........((..((((.......((((((....)))))).....((((...(((((......)))))...))))......)))))).......... (-12.32 = -12.78 + 0.46)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:14:09 2011