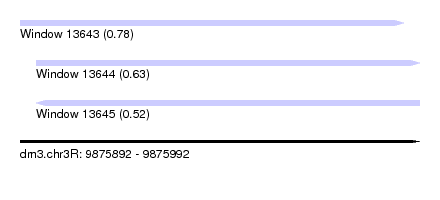
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,875,892 – 9,875,992 |
| Length | 100 |
| Max. P | 0.783314 |

| Location | 9,875,892 – 9,875,988 |
|---|---|
| Length | 96 |
| Sequences | 8 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 81.87 |
| Shannon entropy | 0.31557 |
| G+C content | 0.44263 |
| Mean single sequence MFE | -19.36 |
| Consensus MFE | -17.42 |
| Energy contribution | -17.12 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.783314 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
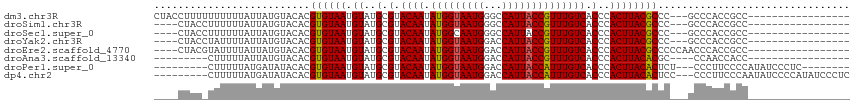
>dm3.chr3R 9875892 96 + 27905053 CUACCUUUUUUUUUUAUUAUGUACACGUGUAAUGUAUGCGUACAAUAUGGUAAUGGGCCAUUACCGUUUGUCACCCACUUACGCCC---GCCCACCGCC----------------- ...................(((((.((((.....)))).)))))....(((..(((((......(((..((.....))..)))...---)))))..)))----------------- ( -19.90, z-score = -0.59, R) >droSim1.chr3R 15963509 92 - 27517382 ----CUACCUUUUUUAUUAUGUACACGUGUAAUGUAUGCGUACAAUAUGGUAAUGGGCCAUUACCGUUUGUCACCCACUUACGCCC---GCCCACCGCC----------------- ----...............(((((.((((.....)))).)))))....(((..(((((......(((..((.....))..)))...---)))))..)))----------------- ( -19.90, z-score = -0.37, R) >droSec1.super_0 8981358 92 - 21120651 ----CUACCUUUUUUAUUAUGUACACGUGUAAUGUAUGCGUACAAUAUGGCAAUGGGCCAUUACCGUUUGUCACCCACUUACGCCC---GCCCACCGCC----------------- ----...............(((((.((((.....)))).)))))....(((..(((((......(((..((.....))..)))...---)))))..)))----------------- ( -21.20, z-score = -0.82, R) >droYak2.chr3R 12275296 92 - 28832112 ----CUACCUAUUUUAUUAUGUACACGUGUAAUGUAUGCGUACAAUAUGGUAAUGGACCAUUACCGUUUGUCACCCACUUACGCCC---GCCCACCGCC----------------- ----......................((((((.((..(.(.((((.(((((((((...)))))))))))))).)..))))))))..---..........----------------- ( -17.10, z-score = 0.13, R) >droEre2.scaffold_4770 9416496 95 + 17746568 ----CUACGUAUUUUAUUAUGUACACGUGUAAUGUAUGCGUACAAUAUGGUAAUGGACCAUUACCGUUUGUCACCCACUUACGCCCCCAACCCACCGCC----------------- ----.((((((......))))))...((((((.((..(.(.((((.(((((((((...)))))))))))))).)..))))))))...............----------------- ( -20.50, z-score = -0.85, R) >droAna3.scaffold_13340 3098738 86 - 23697760 ---------CUUUUUAUUAUGUACACGUGUAAUGUAUGCGUACAAUAUGGUAAUGGACCAUUACCGUUUGUCACCCACUUACACGC----CCAACCACC----------------- ---------................(((((((.((..(.(.((((.(((((((((...)))))))))))))).)..))))))))).----.........----------------- ( -19.70, z-score = -1.33, R) >droPer1.super_0 6930488 96 + 11822988 ---------CUUUUUAUGAUAUACACGUGUAAUGUAUGCGUACAAUAUGGUAAUGGACCAUUACCAUUUGUCACCCACUUACACUCU---CCCUUCCCCAUAUCCCUC-------- ---------.................((((((.((..(.(.((((.(((((((((...)))))))))))))).)..))))))))...---..................-------- ( -18.30, z-score = -2.36, R) >dp4.chr2 17275214 104 - 30794189 ---------CUUUUUAUGAUAUACACGUGUAAUGUAUGCGUACAAUAUGGUAAUGGACCAUUACCAUUUGUCACCCACUUACACUCC---CCCUUCCCAAUAUCCCCAUAUCCCUC ---------.................((((((.((..(.(.((((.(((((((((...)))))))))))))).)..))))))))...---.......................... ( -18.30, z-score = -2.17, R) >consensus ____CUAC_UUUUUUAUUAUGUACACGUGUAAUGUAUGCGUACAAUAUGGUAAUGGACCAUUACCGUUUGUCACCCACUUACGCCC___GCCCACCGCC_________________ ..........................((((((.((..(.(.((((.(((((((((...)))))))))))))).)..))))))))................................ (-17.42 = -17.12 + -0.30)

| Location | 9,875,896 – 9,875,992 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 85.57 |
| Shannon entropy | 0.27447 |
| G+C content | 0.44735 |
| Mean single sequence MFE | -19.80 |
| Consensus MFE | -17.14 |
| Energy contribution | -16.96 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.632459 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
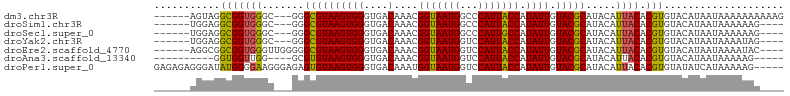
>dm3.chr3R 9875896 96 + 27905053 CUUUUUUUUUUAUUAUGUACACGUGUAAUGUAUGCGUACAAUAUGGUAAUGGGCCAUUACCGUUUGUCACCCACUUACGCCC---GCCCACCGCCUACU------ ...............(((((.((((.....)))).)))))....(((..(((((......(((..((.....))..)))...---)))))..)))....------ ( -21.10, z-score = -1.05, R) >droSim1.chr3R 15963513 92 - 27517382 ----CUUUUUUAUUAUGUACACGUGUAAUGUAUGCGUACAAUAUGGUAAUGGGCCAUUACCGUUUGUCACCCACUUACGCCC---GCCCACCGCCUCCA------ ----...........(((((.((((.....)))).)))))....(((..(((((......(((..((.....))..)))...---)))))..)))....------ ( -21.10, z-score = -0.93, R) >droSec1.super_0 8981362 92 - 21120651 ----CUUUUUUAUUAUGUACACGUGUAAUGUAUGCGUACAAUAUGGCAAUGGGCCAUUACCGUUUGUCACCCACUUACGCCC---GCCCACCGCCUCCA------ ----...........(((((.((((.....)))).)))))....(((..(((((......(((..((.....))..)))...---)))))..)))....------ ( -22.40, z-score = -1.33, R) >droYak2.chr3R 12275300 92 - 28832112 ----CUAUUUUAUUAUGUACACGUGUAAUGUAUGCGUACAAUAUGGUAAUGGACCAUUACCGUUUGUCACCCACUUACGCCC---GCCCACCGCCUCCA------ ----..................((((((.((..(.(.((((.(((((((((...)))))))))))))).)..))))))))..---..............------ ( -17.10, z-score = 0.01, R) >droEre2.scaffold_4770 9416500 95 + 17746568 ----GUAUUUUAUUAUGUACACGUGUAAUGUAUGCGUACAAUAUGGUAAUGGACCAUUACCGUUUGUCACCCACUUACGCCCCCAACCCACCGCCGCCU------ ----((((........))))..((((((.((..(.(.((((.(((((((((...)))))))))))))).)..))))))))...................------ ( -18.90, z-score = -0.32, R) >droAna3.scaffold_13340 3098738 86 - 23697760 -----CUUUUUAUUAUGUACACGUGUAAUGUAUGCGUACAAUAUGGUAAUGGACCAUUACCGUUUGUCACCCACUUACACGC----CCAACCACC---------- -----................(((((((.((..(.(.((((.(((((((((...)))))))))))))).)..))))))))).----.........---------- ( -19.70, z-score = -1.33, R) >droPer1.super_0 6930488 100 + 11822988 -----CUUUUUAUGAUAUACACGUGUAAUGUAUGCGUACAAUAUGGUAAUGGACCAUUACCAUUUGUCACCCACUUACACUCUCCCUUCCCCAUAUCCCUCUCUC -----.................((((((.((..(.(.((((.(((((((((...)))))))))))))).)..))))))))......................... ( -18.30, z-score = -2.62, R) >consensus ____CUUUUUUAUUAUGUACACGUGUAAUGUAUGCGUACAAUAUGGUAAUGGACCAUUACCGUUUGUCACCCACUUACGCCC___GCCCACCGCCUCCA______ ......................((((((.((..(.(.((((.(((((((((...)))))))))))))).)..))))))))......................... (-17.14 = -16.96 + -0.18)

| Location | 9,875,896 – 9,875,992 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 85.57 |
| Shannon entropy | 0.27447 |
| G+C content | 0.44735 |
| Mean single sequence MFE | -26.49 |
| Consensus MFE | -18.46 |
| Energy contribution | -17.97 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.524345 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
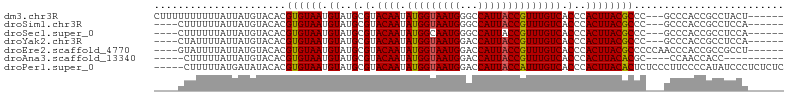
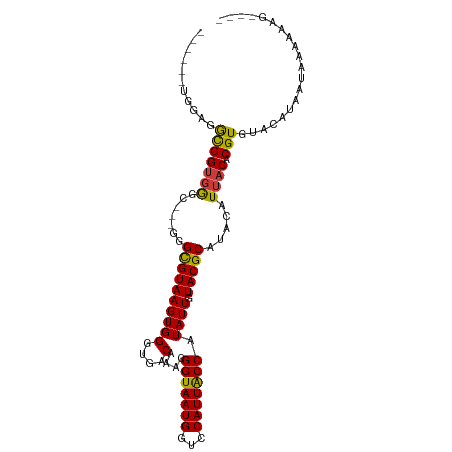
>dm3.chr3R 9875896 96 - 27905053 ------AGUAGGCGGUGGGC---GGGCGUAAGUGGGUGACAAACGGUAAUGGCCCAUUACCAUAUUGUACGCAUACAUUACACGUGUACAUAAUAAAAAAAAAAG ------....((..((((((---..((....)).(....)...........))))))..))....(((((((...........)))))))............... ( -26.80, z-score = -1.52, R) >droSim1.chr3R 15963513 92 + 27517382 ------UGGAGGCGGUGGGC---GGGCGUAAGUGGGUGACAAACGGUAAUGGCCCAUUACCAUAUUGUACGCAUACAUUACACGUGUACAUAAUAAAAAAG---- ------....((..((((((---..((....)).(....)...........))))))..))....(((((((...........)))))))...........---- ( -26.80, z-score = -1.38, R) >droSec1.super_0 8981362 92 + 21120651 ------UGGAGGCGGUGGGC---GGGCGUAAGUGGGUGACAAACGGUAAUGGCCCAUUGCCAUAUUGUACGCAUACAUUACACGUGUACAUAAUAAAAAAG---- ------....((((((((((---..((....)).(....)...........))))))))))....(((((((...........)))))))...........---- ( -31.20, z-score = -2.51, R) >droYak2.chr3R 12275300 92 + 28832112 ------UGGAGGCGGUGGGC---GGGCGUAAGUGGGUGACAAACGGUAAUGGUCCAUUACCAUAUUGUACGCAUACAUUACACGUGUACAUAAUAAAAUAG---- ------....((..((((((---..((....)).(....)...........))))))..))....(((((((...........)))))))...........---- ( -24.10, z-score = -0.73, R) >droEre2.scaffold_4770 9416500 95 - 17746568 ------AGGCGGCGGUGGGUUGGGGGCGUAAGUGGGUGACAAACGGUAAUGGUCCAUUACCAUAUUGUACGCAUACAUUACACGUGUACAUAAUAAAAUAC---- ------....((..(((((..(..(.(((.....(....)..))).)..)..)))))..))....(((((((...........)))))))...........---- ( -24.20, z-score = -0.33, R) >droAna3.scaffold_13340 3098738 86 + 23697760 ----------GGUGGUUGG----GCGUGUAAGUGGGUGACAAACGGUAAUGGUCCAUUACCAUAUUGUACGCAUACAUUACACGUGUACAUAAUAAAAAG----- ----------.(((..((.----(((((((((((.(((((((..(((((((...)))))))...)))).))).))).)))))))).))))).........----- ( -26.10, z-score = -1.94, R) >droPer1.super_0 6930488 100 - 11822988 GAGAGAGGGAUAUGGGGAAGGGAGAGUGUAAGUGGGUGACAAAUGGUAAUGGUCCAUUACCAUAUUGUACGCAUACAUUACACGUGUAUAUCAUAAAAAG----- ........((((((.(.........(((((((((.((((((((((((((((...))))))))).)))).))).))).)))))).).))))))........----- ( -26.20, z-score = -3.05, R) >consensus ______UGGAGGCGGUGGGC___GGGCGUAAGUGGGUGACAAACGGUAAUGGUCCAUUACCAUAUUGUACGCAUACAUUACACGUGUACAUAAUAAAAAAG____ ...........(((((((.......((((((((((....)....(((((((...))))))).)))).))))).....)))).))).................... (-18.46 = -17.97 + -0.48)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:14:07 2011