
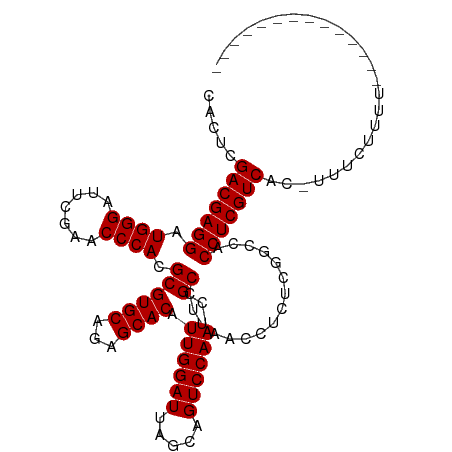
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,870,242 – 9,870,338 |
| Length | 96 |
| Max. P | 0.987009 |

| Location | 9,870,242 – 9,870,338 |
|---|---|
| Length | 96 |
| Sequences | 13 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 90.49 |
| Shannon entropy | 0.21031 |
| G+C content | 0.55062 |
| Mean single sequence MFE | -29.79 |
| Consensus MFE | -29.17 |
| Energy contribution | -29.17 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.98 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.978909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9870242 96 + 27905053 CACUCGACGAGGAUGGGAUUCGAACCCACGCGUGCAGAGCACAUUGGAUUAGCAGUCCAACGCCUUAACCUCUCGGCCACCUCGUCAC-UUUCUUUU-------------- .....(((((((.((((.......)))).((((((...)))).((((((.....)))))).))................)))))))..-........-------------- ( -29.30, z-score = -2.24, R) >droSim1.chr3R 15957910 96 - 27517382 CAUUCGACGAGGAUGGGAUUCGAACCCACGCGUGCAGAGCACAUUGGAUUAGCAGUCCAACGCCUUAACCUCUCGGCCACCUCGUCAC-UUUCUUCU-------------- .....(((((((.((((.......)))).((((((...)))).((((((.....)))))).))................)))))))..-........-------------- ( -29.30, z-score = -2.09, R) >droSec1.super_0 8975748 96 - 21120651 CAUUCGACGAGGAUGGGAUUCGAACCCACGCGUGCAGAGCACAUUGGAUUAGCAGUCCAACGCCUUAACCUCUCGGCCACCUCGUCAC-UUUCUUCU-------------- .....(((((((.((((.......)))).((((((...)))).((((((.....)))))).))................)))))))..-........-------------- ( -29.30, z-score = -2.09, R) >droYak2.chr3R 12269738 96 - 28832112 CACUCGACGAGGAUGGGAUUCGAACCCACGCGUGCAGAGCACAUUGGAUUAGCAGUCCAACGCCUUAACCUCUCGGCCACCUCGUCAC-UUUCUUUU-------------- .....(((((((.((((.......)))).((((((...)))).((((((.....)))))).))................)))))))..-........-------------- ( -29.30, z-score = -2.24, R) >droEre2.scaffold_4770 9411407 96 + 17746568 CACUCGACGAGGAUGGGAUUCGAACCCACGCGUGCAGAGCACAUUGGAUUAGCAGUCCAACGCCUUAACCUCUCGGCCACCUCGUCAC-UUUCUUUU-------------- .....(((((((.((((.......)))).((((((...)))).((((((.....)))))).))................)))))))..-........-------------- ( -29.30, z-score = -2.24, R) >droAna3.scaffold_13340 14778501 86 + 23697760 UAGUCGACGAGGAUGGGAUUCGAACCCACGCGUGCAGAGCACAUUGGAUUAGCAGUCCAACGCCUUAACCACUCGGCCACCUCGUC------------------------- .....(((((((.((((.......)))).((((((...)))).((((((.....)))))).))................)))))))------------------------- ( -27.60, z-score = -1.71, R) >dp4.chr2 17268734 91 - 30794189 ----CGACGAGGAUGGGAUUCGAACCCACGCGUGCAGAGCACAUUGGAUUAGCAGUCCAACGCCUUAACCUCUCGGCCACCUCGUCAC-UUUCUCU--------------- ----.(((((((.((((.......)))).((((((...)))).((((((.....)))))).))................)))))))..-.......--------------- ( -29.30, z-score = -2.06, R) >droPer1.super_0 6924074 93 + 11822988 --AUCGACGAGGAUGGGAUUCGAACCCACGCGUGCAGAGCACAUUGGAUUAGCAGUCCAACGCCUUAACCUCUCGGCCACCUCGUCAC-UUUCUCU--------------- --...(((((((.((((.......)))).((((((...)))).((((((.....)))))).))................)))))))..-.......--------------- ( -29.30, z-score = -2.05, R) >droWil1.scaffold_181108 2024355 96 + 4707319 UCGUCGACGAGGAUGGGAUUCGAACCCACGCGUGCAGAGCACAUUGGAUUAGCAGUCCAACGCCUUAACCUCUCGGCCACCUCGUCAC-UAUAUCCU-------------- .....(((((((.((((.......)))).((((((...)))).((((((.....)))))).))................)))))))..-........-------------- ( -29.30, z-score = -1.59, R) >droVir3.scaffold_12822 3549334 96 - 4096053 CGCUCGACGAGGAUGGGAUUCGAACCCACGCGUGCAGAGCACAUUGGAUUAGCAGUCCAACGCCUUAACCUCUCGGCCACCUCGUCGC-UUUGAACU-------------- ....((((((((.((((.......)))).((((((...)))).((((((.....)))))).))................)))))))).-........-------------- ( -30.20, z-score = -1.18, R) >droMoj3.scaffold_6540 931429 96 - 34148556 CACUCGACGAGGAUGGGAUUCGAACCCACGCGUGCAGAGCACAUUGGAUUAGCAGUCCAACGCCUUAACCUCUCGGCCACCUCGUCGC-UGUGAUUU-------------- (((.((((((((.((((.......)))).((((((...)))).((((((.....)))))).))................)))))))).-.)))....-------------- ( -33.60, z-score = -2.28, R) >droGri2.scaffold_15074 3233009 96 + 7742996 UCGUCGACGAGGAUGGGAUUCGAACCCACGCGUGCAGAGCACAUUGGAUUAGCAGUCCAACGCCUUAACCUCUCGGCCACCUCGUCAC-UUUCUUUC-------------- .....(((((((.((((.......)))).((((((...)))).((((((.....)))))).))................)))))))..-........-------------- ( -29.30, z-score = -1.54, R) >anoGam1.chr2L 39468393 111 + 48795086 UGUUCGACGAGGAUGGGAUUCGAACCCACGCGUGCAGAGCACAUUGGAUUAGCAGUCCAACGCCUUAACCUCUCGGCCACCUCGUCAUGUGUAAGAGUGAAGAGCAACCUA ((..(((.((((.((((.......)))).((((((...)))).((((((.....)))))).)).....)))))))..)).(((.((((........)))).)))....... ( -32.20, z-score = -0.53, R) >consensus CACUCGACGAGGAUGGGAUUCGAACCCACGCGUGCAGAGCACAUUGGAUUAGCAGUCCAACGCCUUAACCUCUCGGCCACCUCGUCAC_UUUCUUUU______________ .....(((((((.((((.......)))).((((((...)))).((((((.....)))))).))................)))))))......................... (-29.17 = -29.17 + -0.00)


| Location | 9,870,242 – 9,870,338 |
|---|---|
| Length | 96 |
| Sequences | 13 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 90.49 |
| Shannon entropy | 0.21031 |
| G+C content | 0.55062 |
| Mean single sequence MFE | -35.68 |
| Consensus MFE | -35.24 |
| Energy contribution | -35.20 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.99 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.987009 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9870242 96 - 27905053 --------------AAAAGAAA-GUGACGAGGUGGCCGAGAGGUUAAGGCGUUGGACUGCUAAUCCAAUGUGCUCUGCACGCGUGGGUUCGAAUCCCAUCCUCGUCGAGUG --------------........-.(((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......))))))))))))).... ( -35.40, z-score = -2.29, R) >droSim1.chr3R 15957910 96 + 27517382 --------------AGAAGAAA-GUGACGAGGUGGCCGAGAGGUUAAGGCGUUGGACUGCUAAUCCAAUGUGCUCUGCACGCGUGGGUUCGAAUCCCAUCCUCGUCGAAUG --------------........-.(((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......))))))))))))).... ( -35.40, z-score = -2.19, R) >droSec1.super_0 8975748 96 + 21120651 --------------AGAAGAAA-GUGACGAGGUGGCCGAGAGGUUAAGGCGUUGGACUGCUAAUCCAAUGUGCUCUGCACGCGUGGGUUCGAAUCCCAUCCUCGUCGAAUG --------------........-.(((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......))))))))))))).... ( -35.40, z-score = -2.19, R) >droYak2.chr3R 12269738 96 + 28832112 --------------AAAAGAAA-GUGACGAGGUGGCCGAGAGGUUAAGGCGUUGGACUGCUAAUCCAAUGUGCUCUGCACGCGUGGGUUCGAAUCCCAUCCUCGUCGAGUG --------------........-.(((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......))))))))))))).... ( -35.40, z-score = -2.29, R) >droEre2.scaffold_4770 9411407 96 - 17746568 --------------AAAAGAAA-GUGACGAGGUGGCCGAGAGGUUAAGGCGUUGGACUGCUAAUCCAAUGUGCUCUGCACGCGUGGGUUCGAAUCCCAUCCUCGUCGAGUG --------------........-.(((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......))))))))))))).... ( -35.40, z-score = -2.29, R) >droAna3.scaffold_13340 14778501 86 - 23697760 -------------------------GACGAGGUGGCCGAGUGGUUAAGGCGUUGGACUGCUAAUCCAAUGUGCUCUGCACGCGUGGGUUCGAAUCCCAUCCUCGUCGACUA -------------------------(((((((((((.((((.......((((((((.......)))))))))))).)).)))(((((.......))))).))))))..... ( -33.51, z-score = -1.95, R) >dp4.chr2 17268734 91 + 30794189 ---------------AGAGAAA-GUGACGAGGUGGCCGAGAGGUUAAGGCGUUGGACUGCUAAUCCAAUGUGCUCUGCACGCGUGGGUUCGAAUCCCAUCCUCGUCG---- ---------------.......-..((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......)))))))))))).---- ( -35.20, z-score = -2.22, R) >droPer1.super_0 6924074 93 - 11822988 ---------------AGAGAAA-GUGACGAGGUGGCCGAGAGGUUAAGGCGUUGGACUGCUAAUCCAAUGUGCUCUGCACGCGUGGGUUCGAAUCCCAUCCUCGUCGAU-- ---------------.......-.(((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......)))))))))))))..-- ( -35.40, z-score = -2.12, R) >droWil1.scaffold_181108 2024355 96 - 4707319 --------------AGGAUAUA-GUGACGAGGUGGCCGAGAGGUUAAGGCGUUGGACUGCUAAUCCAAUGUGCUCUGCACGCGUGGGUUCGAAUCCCAUCCUCGUCGACGA --------------........-((((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......)))))))))))).)).. ( -35.90, z-score = -1.54, R) >droVir3.scaffold_12822 3549334 96 + 4096053 --------------AGUUCAAA-GCGACGAGGUGGCCGAGAGGUUAAGGCGUUGGACUGCUAAUCCAAUGUGCUCUGCACGCGUGGGUUCGAAUCCCAUCCUCGUCGAGCG --------------........-.(((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......))))))))))))).... ( -37.20, z-score = -1.82, R) >droMoj3.scaffold_6540 931429 96 + 34148556 --------------AAAUCACA-GCGACGAGGUGGCCGAGAGGUUAAGGCGUUGGACUGCUAAUCCAAUGUGCUCUGCACGCGUGGGUUCGAAUCCCAUCCUCGUCGAGUG --------------....(((.-.(((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......))))))))))))).))) ( -38.20, z-score = -2.40, R) >droGri2.scaffold_15074 3233009 96 - 7742996 --------------GAAAGAAA-GUGACGAGGUGGCCGAGAGGUUAAGGCGUUGGACUGCUAAUCCAAUGUGCUCUGCACGCGUGGGUUCGAAUCCCAUCCUCGUCGACGA --------------........-((((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......)))))))))))).)).. ( -35.90, z-score = -1.90, R) >anoGam1.chr2L 39468393 111 - 48795086 UAGGUUGCUCUUCACUCUUACACAUGACGAGGUGGCCGAGAGGUUAAGGCGUUGGACUGCUAAUCCAAUGUGCUCUGCACGCGUGGGUUCGAAUCCCAUCCUCGUCGAACA ........................(((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......))))))))))))).... ( -35.50, z-score = -0.57, R) >consensus ______________AAAAGAAA_GUGACGAGGUGGCCGAGAGGUUAAGGCGUUGGACUGCUAAUCCAAUGUGCUCUGCACGCGUGGGUUCGAAUCCCAUCCUCGUCGAGUG ........................(((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......))))))))))))).... (-35.24 = -35.20 + -0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:14:05 2011