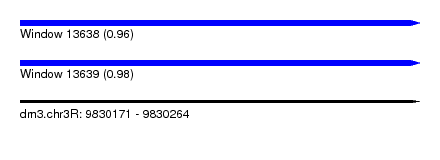
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,830,171 – 9,830,264 |
| Length | 93 |
| Max. P | 0.975388 |

| Location | 9,830,171 – 9,830,264 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 86.10 |
| Shannon entropy | 0.20373 |
| G+C content | 0.35614 |
| Mean single sequence MFE | -20.29 |
| Consensus MFE | -15.15 |
| Energy contribution | -15.15 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.956213 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
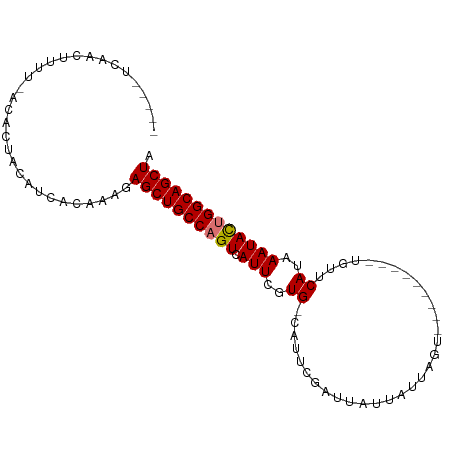
>dm3.chr3R 9830171 93 + 27905053 -----UCAACUUUUUACACUACAUCACAAAGAGCUGCCAGUCAUUCGUG-CAUUUGAUUAUUAUUAUUAUUAUUAGUUGUUCAAAAAUAUUGGCAGCUA -----..........................((((((((((.......(-((.(((((...((....))..))))).)))........)))))))))). ( -18.36, z-score = -2.27, R) >droSim1.chr3R 15918562 88 - 27517382 -----UCAACUUUUUACACUACAUCACAAAGAGCUGCCAGUCAUUCGUGGCAUUCGAUUAUUAUUAUUA------GUUGUUCAUAAAUAUUGGCAGCUA -----..........................((((((((((.(((..(((....((((((.......))------)))).)))..))))))))))))). ( -22.20, z-score = -4.01, R) >droYak2.chr3R 12230861 88 - 28832112 GCCGAUCAACUUUU-GCACUACAUCACAAAGAGCUGCCAGUCAUUCGUG-CAUUCGAUUAUUAUUUGU---------UGUUCAUAAAUACGGGCAGCUA ((.((......)).-))..............(((((((.((.(((..((-....((((........))---------))..))..))))).))))))). ( -18.50, z-score = -0.95, R) >droEre2.scaffold_4770 9373714 83 + 17746568 -----UCAACUUUU-GCACUACAUCACAAAGAGCUGCCAGUCAUUCGUG-CAUUCGAUUAUUAUUUGU---------UGUUCAUAAAUACUGGCAGCUA -----.........-................((((((((((.(((..((-....((((........))---------))..))..))))))))))))). ( -22.10, z-score = -3.24, R) >consensus _____UCAACUUUU_ACACUACAUCACAAAGAGCUGCCAGUCAUUCGUG_CAUUCGAUUAUUAUUAGU_________UGUUCAUAAAUACUGGCAGCUA ...............................((((((((((.(((..((................................))..))))))))))))). (-15.15 = -15.15 + 0.00)

| Location | 9,830,171 – 9,830,264 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 88.26 |
| Shannon entropy | 0.17433 |
| G+C content | 0.35093 |
| Mean single sequence MFE | -20.04 |
| Consensus MFE | -16.37 |
| Energy contribution | -16.18 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.975388 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
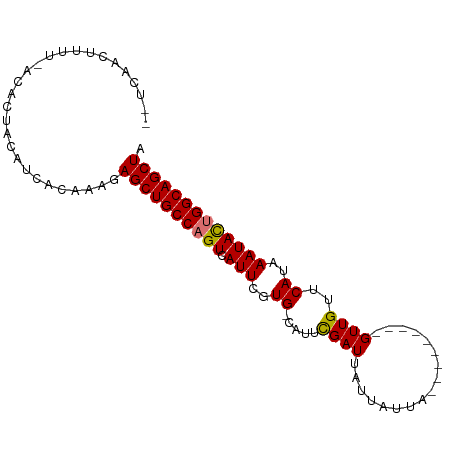
>dm3.chr3R 9830171 93 + 27905053 --UCAACUUUUUACACUACAUCACAAAGAGCUGCCAGUCAUUCGUG-CAUUUGAUUAUUAUUAUUAUUAUUAGUUGUUCAAAAAUAUUGGCAGCUA --..........................((((((((((.......(-((.(((((...((....))..))))).)))........)))))))))). ( -18.36, z-score = -2.27, R) >droSim1.chr3R 15918562 88 - 27517382 --UCAACUUUUUACACUACAUCACAAAGAGCUGCCAGUCAUUCGUGGCAUUCGAUUAUUAUUAUUA------GUUGUUCAUAAAUAUUGGCAGCUA --..........................((((((((((.(((..(((....((((((.......))------)))).)))..))))))))))))). ( -22.20, z-score = -4.01, R) >droYak2.chr3R 12230864 85 - 28832112 GAUCAACUUUU-GCACUACAUCACAAAGAGCUGCCAGUCAUUCGUG-CAUUCGAUUAUUAUUU---------GUUGUUCAUAAAUACGGGCAGCUA ...........-................(((((((.((.(((..((-....((((........---------))))..))..))))).))))))). ( -17.50, z-score = -1.09, R) >droEre2.scaffold_4770 9373714 83 + 17746568 --UCAACUUUU-GCACUACAUCACAAAGAGCUGCCAGUCAUUCGUG-CAUUCGAUUAUUAUUU---------GUUGUUCAUAAAUACUGGCAGCUA --.........-................((((((((((.(((..((-....((((........---------))))..))..))))))))))))). ( -22.10, z-score = -3.24, R) >consensus __UCAACUUUU_ACACUACAUCACAAAGAGCUGCCAGUCAUUCGUG_CAUUCGAUUAUUAUUA_________GUUGUUCAUAAAUACUGGCAGCUA ............................((((((((((.(((..((.....((((.................))))..))..))))))))))))). (-16.37 = -16.18 + -0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:14:02 2011