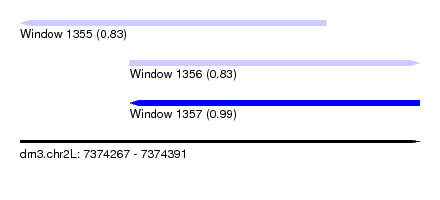
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,374,267 – 7,374,391 |
| Length | 124 |
| Max. P | 0.991221 |

| Location | 7,374,267 – 7,374,362 |
|---|---|
| Length | 95 |
| Sequences | 12 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 85.25 |
| Shannon entropy | 0.29690 |
| G+C content | 0.46467 |
| Mean single sequence MFE | -33.60 |
| Consensus MFE | -15.17 |
| Energy contribution | -14.79 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.831113 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
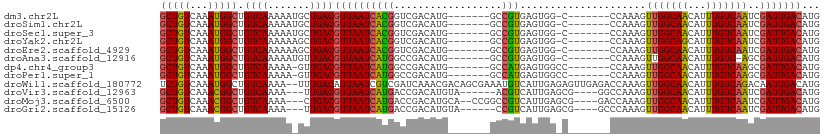
>dm3.chr2L 7374267 95 - 23011544 GCUGUCAAAUGGCUGUCAAAAAUGCUGACGUUAAUCACGGUCGACAUG-------GCCGUGAGUGG-C-------CCAAAGUUGGCAACAUUUGUCAAUCGAUUGACAUG ..((((((.((..((.((((..(((..((((((.((((((((.....)-------))))))).)))-)-------.....))..)))...)))).))..)).)))))).. ( -34.80, z-score = -3.21, R) >droSim1.chr2L 7162396 95 - 22036055 GCUGUCAAAUGGCUGUCAAAAAUGCUGACGUUAAUCACGGUCGACAUG-------GCCGUGAGUGG-C-------CCAAAGUUGGCAACAUUUGUCAAUCGAUUGACAUG ..((((((.((..((.((((..(((..((((((.((((((((.....)-------))))))).)))-)-------.....))..)))...)))).))..)).)))))).. ( -34.80, z-score = -3.21, R) >droSec1.super_3 2889296 95 - 7220098 GCUGUCAAAUGGCUGUCAAAAAUGCUGACGUUAAUCACGGUCGACAUG-------GCCGUGAGUGG-C-------CCAAAGUUGGCAACAUUUGUCAAUCGAUUGACAUG ..((((((.((..((.((((..(((..((((((.((((((((.....)-------))))))).)))-)-------.....))..)))...)))).))..)).)))))).. ( -34.80, z-score = -3.21, R) >droYak2.chr2L 16794962 95 + 22324452 GCUGUCAAAUGGCUGUCAAAAAAGCUGACGUUAAUCACGGUCGACAUG-------GCCGUGAGUGG-C-------CCAAAGUUGGCAGCAUUUGUCAAUCGAUUGACAUG ((((((((.(((..((((.......))))((((.((((((((.....)-------))))))).)))-)-------)))...))))))))...((((((....)))))).. ( -37.80, z-score = -4.06, R) >droEre2.scaffold_4929 16289301 95 - 26641161 GCUGUCAAAUGGCUGUCAAAAAAGCUGACGUUAAUCACGGUCGACAUG-------GCCGUGAGUGG-C-------CCAAAGUUGGCAACAUUUGUCAAUCGAUUGACAUG ..((((((.((..((.((((...((..((((((.((((((((.....)-------))))))).)))-)-------.....))..))....)))).))..)).)))))).. ( -33.00, z-score = -2.83, R) >droAna3.scaffold_12916 15279398 94 - 16180835 GCUGUCAAAUGGCUGUCAAAAAUGUUGACGUUAAUCAUGGCCGACAUG-------GCCGUGAGUGG-C-------CCAAAGUUGGCAACAUUUGUC-AGCGAUUGACAUG ((((.((((((..((((((.....(((..((((.((((((((.....)-------))))))).)))-)-------.)))..)))))).)))))).)-))).......... ( -37.80, z-score = -3.83, R) >dp4.chr4_group3 10979702 95 + 11692001 GCUGUCAAAUGGCUGUCAAAAA-GUUGACGUUAAUCAUGGCCGACAUG-------GCCAUGAGUGGCC-------CCAAAGUUGGCAACAUUUGUCAAGCGAUUGACAUG ((((.((((((..((((((...-.(((..((((.((((((((.....)-------))))))).)))).-------.)))..)))))).)))))).).))).......... ( -35.40, z-score = -3.32, R) >droPer1.super_1 8094323 95 + 10282868 GCUGUCAAAUGGCUGUCAAAAA-GUUGACGUUAAUCAUGGCCGACAUG-------GCCAUGAGUGGCC-------CCAAAGUUGGCAACAUUUGUCAAGCGAUUGACAUG ((((.((((((..((((((...-.(((..((((.((((((((.....)-------))))))).)))).-------.)))..)))))).)))))).).))).......... ( -35.40, z-score = -3.32, R) >droWil1.scaffold_180772 3571521 108 + 8906247 UCUGUCAAAUGGCUGUCAAAA--UUUGACAUUAAUCGUCGAUCAAACGACAGCGAAAUGUCAUUGAGAGUUGAGACCAAAGUUGGCAACAUUUGUCAGACAAUUGACAUG ..((((((.((.(((.((((.--..(((((((..((((.(.((....))).)))))))))))......((((.(((....)))..)))).)))).))).)).)))))).. ( -29.70, z-score = -1.83, R) >droVir3.scaffold_12963 4714248 97 + 20206255 GCUGUCAAACGGCUGUCAAAA---UUGACGUUAAUCAUGACCGACAUGUA------ACGUCAUUGAGCG----GGCCAAAGUUGGCAACAUUUGUCAAUCGAUUGACAUG ..((((((.((..((.((((.---.((((((((..((((.....))))))------)))))).......----.((((....))))....)))).))..)).)))))).. ( -31.20, z-score = -2.57, R) >droMoj3.scaffold_6500 13879507 101 + 32352404 GCUGUCAAACGGCUGUCAAAA---CUGACGUUAAUCAUGACCGACAUGCA--CCGGCCGUCAUUGAGCG----GACCAAAGUUGGCAACAUUUGUCAAUCGAUUGACAUG ((((.....)))).((((...---.))))(((((((..............--..((((((......)))----).))...(((((((.....))))))).)))))))... ( -29.00, z-score = -1.18, R) >droGri2.scaffold_15126 4947068 97 - 8399593 GCUGUCAAACGGCUGUCAAAA---UUGACGUUAAUCAUGACCGACAUGUA------CCGUCAUUGAGCG----GCCCAAAGUUGGCAACAUUUGUCAAUCGAUUGACAUG ..((((((.((((((((((..---.(((((.....((((.....))))..------.))))))))).))----)))....(((((((.....))))))).).)))))).. ( -29.50, z-score = -2.16, R) >consensus GCUGUCAAAUGGCUGUCAAAAA_GCUGACGUUAAUCAUGGCCGACAUG_______GCCGUGAGUGG_C_______CCAAAGUUGGCAACAUUUGUCAAUCGAUUGACAUG (((((...))))).((((.......))))((((((((((..................))).....................(((((((...)))))))..)))))))... (-15.17 = -14.79 + -0.38)
| Location | 7,374,301 – 7,374,391 |
|---|---|
| Length | 90 |
| Sequences | 10 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 82.30 |
| Shannon entropy | 0.33477 |
| G+C content | 0.47908 |
| Mean single sequence MFE | -28.21 |
| Consensus MFE | -15.37 |
| Energy contribution | -15.81 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.833099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 7374301 90 + 23011544 ----GGCCACUCACGGC-CAUGUCGACCGUGAUUAACGUCAGCAUUUUUGACAGCCAUUUGACAGCUGUC------AGCGCUGCAUAAGGCAUUUCAAAUA ----((((......)))-)((((((((.((.....))))).(((.(.((((((((.........))))))------)).).)))....)))))........ ( -28.90, z-score = -2.58, R) >droSim1.chr2L 7162430 90 + 22036055 ----GGCCACUCACGGC-CAUGUCGACCGUGAUUAACGUCAGCAUUUUUGACAGCCAUUUGACAGCUGUC------AGCGCUGCAUAAGGCAUUUCAAAUA ----((((......)))-)((((((((.((.....))))).(((.(.((((((((.........))))))------)).).)))....)))))........ ( -28.90, z-score = -2.58, R) >droSec1.super_3 2889330 90 + 7220098 ----GGCCACUCACGGC-CAUGUCGACCGUGAUUAACGUCAGCAUUUUUGACAGCCAUUUGACAGCUGUC------AGCGCUGCAUAAGGCAUUUCAAAUA ----((((......)))-)((((((((.((.....))))).(((.(.((((((((.........))))))------)).).)))....)))))........ ( -28.90, z-score = -2.58, R) >droYak2.chr2L 16794996 90 - 22324452 ----GGCCACUCACGGC-CAUGUCGACCGUGAUUAACGUCAGCUUUUUUGACAGCCAUUUGACAGCUGUC------AGCGCUGCAUAAGGCAUUUCAAAAA ----.(((..((((((.-(.....).)))))).....(.((((....((((((((.........))))))------)).)))))....))).......... ( -28.50, z-score = -2.30, R) >droEre2.scaffold_4929 16289335 90 + 26641161 ----GGCCACUCACGGC-CAUGUCGACCGUGAUUAACGUCAGCUUUUUUGACAGCCAUUUGACAGCUGUC------AGCGCUGCAUAAGGCAUUUCAAAAA ----.(((..((((((.-(.....).)))))).....(.((((....((((((((.........))))))------)).)))))....))).......... ( -28.50, z-score = -2.30, R) >droAna3.scaffold_12916 15279431 90 + 16180835 ----GGCCACUCACGGC-CAUGUCGGCCAUGAUUAACGUCAACAUUUUUGACAGCCAUUUGACAGCUGCC------ACCGCUGCAUAAGGCAUUUCCACAA ----((((......)))-).(((.((...........(((((.....))))).(((...((.((((....------...))))))...)))....))))). ( -25.80, z-score = -2.04, R) >dp4.chr4_group3 10979737 95 - 11692001 ----GGCCACUCAUGGC-CAUGUCGGCCAUGAUUAACGUCAAC-UUUUUGACAGCCAUUUGACAGCUGCUGUUGCUAUUGCUGCAUAAGGCAUUUAAAAUA ----......(((((((-(.....)))))))).....(((((.-...))))).(((...((.((((.((....))....))))))...))).......... ( -31.80, z-score = -2.83, R) >droPer1.super_1 8094358 95 - 10282868 ----GGCCACUCAUGGC-CAUGUCGGCCAUGAUUAACGUCAAC-UUUUUGACAGCCAUUUGACAGCUGCUGUUGCUAUUGCUGCAUAAGGCAUUUCAAAUA ----(((...(((((((-(.....)))))))).....(((((.-...))))).)))((((((..(((..(((.((....)).)))...)))...)))))). ( -33.90, z-score = -3.43, R) >droVir3.scaffold_12963 4714282 84 - 20206255 GCCCGCUCAAUGACGUUACAUGUCGGUCAUGAUUAACGUCAA---UUUUGACAGCCGUUUGACAGCUGUU------------GCAUAAGCCGU--CAACUA ....((((((((((((((((((.....))))..)))))))).---..))))..))...(((((.(((...------------.....))).))--)))... ( -22.50, z-score = -1.51, R) >droGri2.scaffold_15126 4947102 84 + 8399593 GGCCGCUCAAUGACGGUACAUGUCGGUCAUGAUUAACGUCAA---UUUUGACAGCCGUUUGACAGCUGUU------------GCAUAAGCCGU--CAACUA (((.(.(((((((((...((((.....)))).....))))).---..))))).)))..(((((.(((...------------.....))).))--)))... ( -24.40, z-score = -1.31, R) >consensus ____GGCCACUCACGGC_CAUGUCGACCAUGAUUAACGUCAAC_UUUUUGACAGCCAUUUGACAGCUGUC______AGCGCUGCAUAAGGCAUUUCAAAUA .....(((......(((.((((.....))))......(((((.....))))).)))...((.((((.............))))))...))).......... (-15.37 = -15.81 + 0.44)
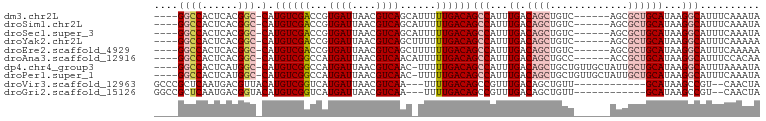
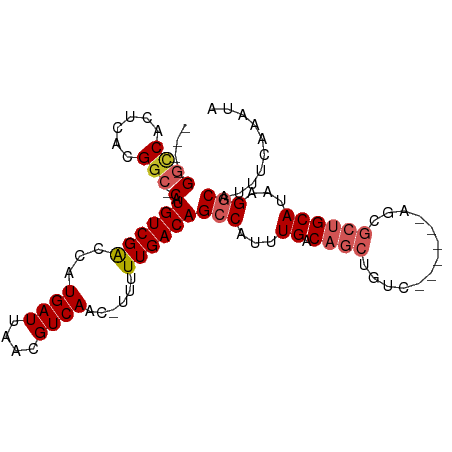
| Location | 7,374,301 – 7,374,391 |
|---|---|
| Length | 90 |
| Sequences | 10 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 82.30 |
| Shannon entropy | 0.33477 |
| G+C content | 0.47908 |
| Mean single sequence MFE | -32.77 |
| Consensus MFE | -19.37 |
| Energy contribution | -19.74 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.991221 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
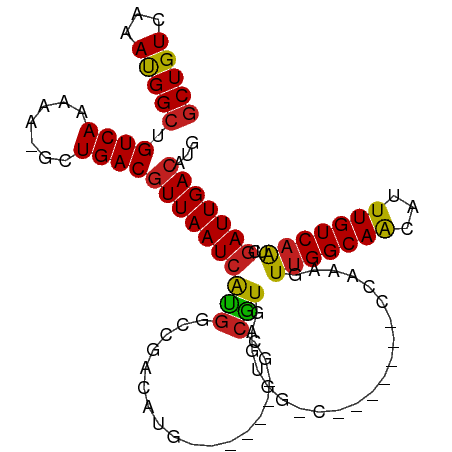
>dm3.chr2L 7374301 90 - 23011544 UAUUUGAAAUGCCUUAUGCAGCGCU------GACAGCUGUCAAAUGGCUGUCAAAAAUGCUGACGUUAAUCACGGUCGACAUG-GCCGUGAGUGGCC---- ..........(((..((((((((.(------(((((((((...))))))))))....))))).)))...((((((((.....)-)))))))..))).---- ( -34.40, z-score = -3.52, R) >droSim1.chr2L 7162430 90 - 22036055 UAUUUGAAAUGCCUUAUGCAGCGCU------GACAGCUGUCAAAUGGCUGUCAAAAAUGCUGACGUUAAUCACGGUCGACAUG-GCCGUGAGUGGCC---- ..........(((..((((((((.(------(((((((((...))))))))))....))))).)))...((((((((.....)-)))))))..))).---- ( -34.40, z-score = -3.52, R) >droSec1.super_3 2889330 90 - 7220098 UAUUUGAAAUGCCUUAUGCAGCGCU------GACAGCUGUCAAAUGGCUGUCAAAAAUGCUGACGUUAAUCACGGUCGACAUG-GCCGUGAGUGGCC---- ..........(((..((((((((.(------(((((((((...))))))))))....))))).)))...((((((((.....)-)))))))..))).---- ( -34.40, z-score = -3.52, R) >droYak2.chr2L 16794996 90 + 22324452 UUUUUGAAAUGCCUUAUGCAGCGCU------GACAGCUGUCAAAUGGCUGUCAAAAAAGCUGACGUUAAUCACGGUCGACAUG-GCCGUGAGUGGCC---- ..........(((..(((((((..(------(((((((((...)))))))))).....)))).)))...((((((((.....)-)))))))..))).---- ( -34.40, z-score = -3.42, R) >droEre2.scaffold_4929 16289335 90 - 26641161 UUUUUGAAAUGCCUUAUGCAGCGCU------GACAGCUGUCAAAUGGCUGUCAAAAAAGCUGACGUUAAUCACGGUCGACAUG-GCCGUGAGUGGCC---- ..........(((..(((((((..(------(((((((((...)))))))))).....)))).)))...((((((((.....)-)))))))..))).---- ( -34.40, z-score = -3.42, R) >droAna3.scaffold_12916 15279431 90 - 16180835 UUGUGGAAAUGCCUUAUGCAGCGGU------GGCAGCUGUCAAAUGGCUGUCAAAAAUGUUGACGUUAAUCAUGGCCGACAUG-GCCGUGAGUGGCC---- ..........(((..((((((((.(------(((((((((...))))))))))....))))).)))...((((((((.....)-)))))))..))).---- ( -34.50, z-score = -2.56, R) >dp4.chr4_group3 10979737 95 + 11692001 UAUUUUAAAUGCCUUAUGCAGCAAUAGCAACAGCAGCUGUCAAAUGGCUGUCAAAAA-GUUGACGUUAAUCAUGGCCGACAUG-GCCAUGAGUGGCC---- ..........(((...((((((....((....)).)))).))...))).(((((...-.)))))((((.((((((((.....)-))))))).)))).---- ( -31.80, z-score = -2.57, R) >droPer1.super_1 8094358 95 + 10282868 UAUUUGAAAUGCCUUAUGCAGCAAUAGCAACAGCAGCUGUCAAAUGGCUGUCAAAAA-GUUGACGUUAAUCAUGGCCGACAUG-GCCAUGAGUGGCC---- ..(((((...(((...((((((....((....)).)))).))...)))..)))))..-......((((.((((((((.....)-))))))).)))).---- ( -32.30, z-score = -2.53, R) >droVir3.scaffold_12963 4714282 84 + 20206255 UAGUUG--ACGGCUUAUGC------------AACAGCUGUCAAACGGCUGUCAAAA---UUGACGUUAAUCAUGACCGACAUGUAACGUCAUUGAGCGGGC ...(((--((((((.....------------...)))))))))..(.(((((((..---.((((((((..((((.....)))))))))))))))).))).) ( -28.10, z-score = -2.81, R) >droGri2.scaffold_15126 4947102 84 - 8399593 UAGUUG--ACGGCUUAUGC------------AACAGCUGUCAAACGGCUGUCAAAA---UUGACGUUAAUCAUGACCGACAUGUACCGUCAUUGAGCGGCC ...(((--((((((.....------------...)))))))))..(((((((((..---.(((((.....((((.....))))...))))))))).))))) ( -29.00, z-score = -3.08, R) >consensus UAUUUGAAAUGCCUUAUGCAGCGCU______GACAGCUGUCAAAUGGCUGUCAAAAA_GCUGACGUUAAUCACGGCCGACAUG_GCCGUGAGUGGCC____ ..........(((..(((((((..........((((((((...)))))))).......)))).)))...(((((((........)))))))..)))..... (-19.37 = -19.74 + 0.37)
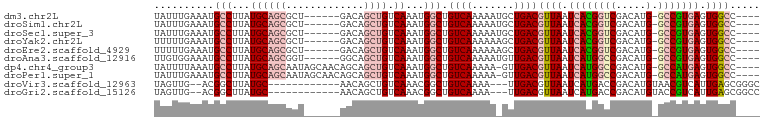
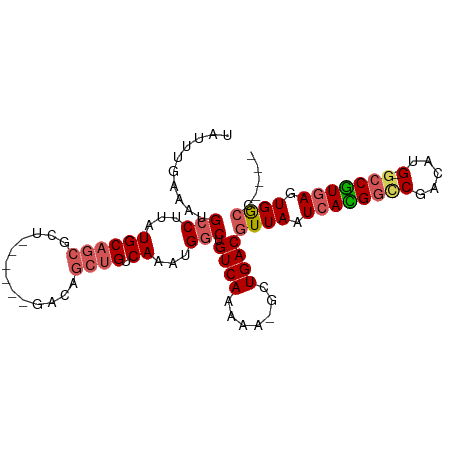
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:23:35 2011