
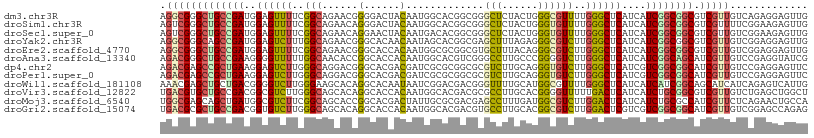
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,791,480 – 9,791,588 |
| Length | 108 |
| Max. P | 0.559750 |

| Location | 9,791,480 – 9,791,588 |
|---|---|
| Length | 108 |
| Sequences | 12 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 77.46 |
| Shannon entropy | 0.50214 |
| G+C content | 0.61420 |
| Mean single sequence MFE | -41.59 |
| Consensus MFE | -25.19 |
| Energy contribution | -24.73 |
| Covariance contribution | -0.46 |
| Combinations/Pair | 1.54 |
| Mean z-score | -0.88 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.559750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
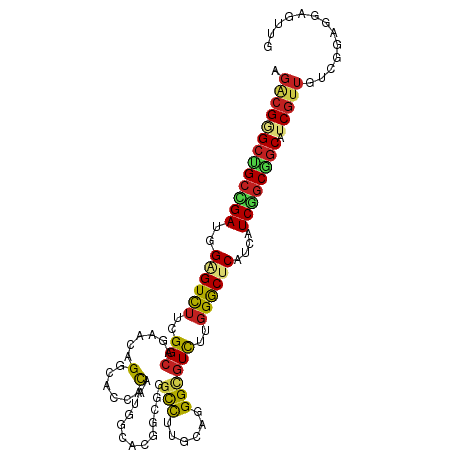
>dm3.chr3R 9791480 108 - 27905053 AGGCGGGCUGCCGAUGGAGUUUUCGGCAGAACGGGGACUACAAUGGCACGGCGGGCUCUACUGGGCGUUUUGGGCUCAUCAUCGGCGGCGUCGUUGUCAGAGGAGUUG .((((((((((((((((((((..((......))..))))....((((.(((.(.((((....)))).).))).)).))))))))))))).)))))............. ( -43.50, z-score = -2.06, R) >droSim1.chr3R 15881525 108 + 27517382 AGUCGGGCUGCCGAUGGAGUUUUCGGCAGAACAGGGACUACAAUGGCACGGCGGGCUCUACUGGGUGUUUUGGGCUCAUCAUCGGCGGCGUCGUUUUCGGAAGAGUUG ...(((((((((((((((((((..(......)..))))).............((((((.((.....))...)))))).))))))))))).)))..(((....)))... ( -38.20, z-score = -1.13, R) >droSec1.super_0 8902028 108 + 21120651 AGUCGGGCUGCCGAUGGAGUUUUCGGCAGAACAGGAACUACAAUGACACGGCGGGCUCUACUGGGUGUUUUGGGCUCAUCAUCGGCGGCGUCGUUGUCGGAAGAGUUG ...(((((((((((((((((((..(......)..))))).............((((((.((.....))...)))))).))))))))))).)))...((....)).... ( -38.20, z-score = -1.10, R) >droYak2.chr3R 12194060 108 + 28832112 AGGCGGGCAGCCGAUGGAGUCUUUGGCAGAACGGGCACAACAAUAGCACGGCGAGCUUUAGAGGGCGUCUUGGGCUCAUCAUCGGCGGCGUCGUUGUCGGAGGAGUUG .(((((((.(((((((((((((..(((....((.((.........)).))....((((....)))))))..))))))..))))))).)).)))))............. ( -37.50, z-score = -0.55, R) >droEre2.scaffold_4770 9337121 108 - 17746568 AGGCGGGCUGCCGAUGGAGUUUUCGGCAGAACGGGCACCACAAUGGCGCGGCGUGCUUUACAGGGCGUCUUGGGCUCAUCAUCGGCGGCGUCGUUGUCGGAGGAGUUG ...((..(((((((........)))))))..))....(((((((((((((((((.((....)).)))))....(((.......))).)))))))))).))........ ( -44.50, z-score = -1.25, R) >droAna3.scaffold_13340 3019244 108 + 23697760 AGACGGGCUGCCGAAGGGGUUUUUGGCAACACCGGCACCACAAUGGCACGUCGGGCCUUGCCCGGGGUCUUGGGCUCAUCAUCGGCAGCAUCGUUGUCCGAGGUAUCG .(((((((((((((.(((((...((....))(((((.((.....))...))))))))))((((((....))))))......)))))))).)))))...(((....))) ( -44.70, z-score = -1.28, R) >dp4.chr2 17181218 108 + 30794189 AGACGAGCCGCUGAAGGAGUCUUGGGCAGGACGGGCACGACGAUCGCGCGGCGCGUCUUGCAGGGUGUCUUGGGCUCAUCGUCGGCGGCAUCGUUGUCCGAGGAGUUC .(((((((((((((..((((((.((((((((((.((.((.((....)))))).)))))))).......)).))))))....)))))))).))))).((....)).... ( -50.51, z-score = -1.75, R) >droPer1.super_0 6834650 108 - 11822988 AGACGAGCCGCUGAAGGAGUCUUGGGCAGGACGGGCACGACGAUCGCGCGGCGCGUCUUGCAGGGUGUCUUGGGCUCAUCGUCGGCGGCAUCGUUGUCCGAGGAGUUC .(((((((((((((..((((((.((((((((((.((.((.((....)))))).)))))))).......)).))))))....)))))))).))))).((....)).... ( -50.51, z-score = -1.75, R) >droWil1.scaffold_181108 1926952 108 - 4707319 AAACGAGCUGCUGACGGGGUCUUGGGAAGCACAGGCACAAUAAUCGGACGACGGGUUUUGCAUGGCGUUUUGGGCUCAUCAUCAUCGGCAGCAUCAUCAGAGUCAUUG ....((((((((((..(((.((..(((.((.((.(((.(((..(((.....)))))).))).)))).)))..))))).......)))))))).))............. ( -30.90, z-score = -0.88, R) >droVir3.scaffold_12822 3465221 108 + 4096053 UGACGUGCUGCCGACGGCGUCUUGGGCAGCACAGGCACCACAAUGGCACGACGCGCCUUGCACGGGGUUUUUGACUCAUCAUCUGCGGCGUCGUUGUCUGAGCUGGCU .(((..((((....)))))))..((.((((.((((((((.....)).(((((((((..((....(((((...)))))..))...)).))))))))))))).)))).)) ( -38.50, z-score = 0.89, R) >droMoj3.scaffold_6540 863163 108 + 34148556 UGGCGAGCAGCUGAUGGCGUCUUCGGCAGCACCGGCACGACUAUUGCGCGACGAGCCUUUGAUGGCGUCUUGGACUCAUCAUCUGCGCCAUCGUUCUCAGAACUGCCA .((((.((((.((((((.((((..(((.....(((((.......))).))....(((......))))))..)))))))))).))))......((((...)))))))). ( -38.00, z-score = 0.02, R) >droGri2.scaffold_15074 3156339 108 - 7742996 UGACGCGCUGCCGACGGUGUCUUGGGCAGCACAGGCACCACAAUGGCACGACGUGCCUUGCACGGCGUCUUGGACUCGUCGUCGGCGGCAUCGUUGUCGGAGCCAGAG ....(((((((((((((.((((.((((.((....(((.......(((((...))))).)))...)))))).))))...))))))))))).(((....))).))..... ( -44.10, z-score = 0.26, R) >consensus AGACGGGCUGCCGAUGGAGUCUUCGGCAGAACAGGCACCACAAUGGCACGGCGGGCCUUGCAGGGCGUCUUGGGCUCAUCAUCGGCGGCAUCGUUGUCGGAGGAGUUG .(((((((((((((..((((((..(((......(......).............(((......))))))..))))))....)))))))).)))))............. (-25.19 = -24.73 + -0.46)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:13:52 2011