
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,766,701 – 9,766,828 |
| Length | 127 |
| Max. P | 0.635687 |

| Location | 9,766,701 – 9,766,796 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 87.08 |
| Shannon entropy | 0.22227 |
| G+C content | 0.55612 |
| Mean single sequence MFE | -34.48 |
| Consensus MFE | -29.56 |
| Energy contribution | -30.82 |
| Covariance contribution | 1.26 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.635687 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
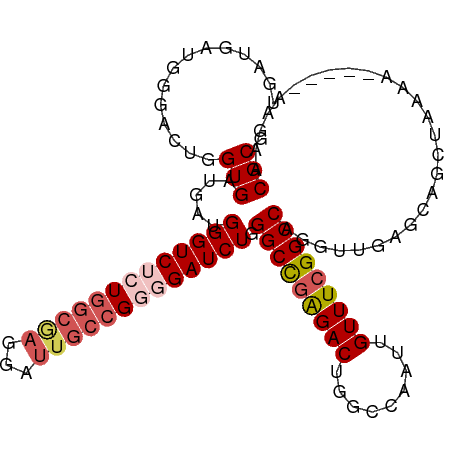
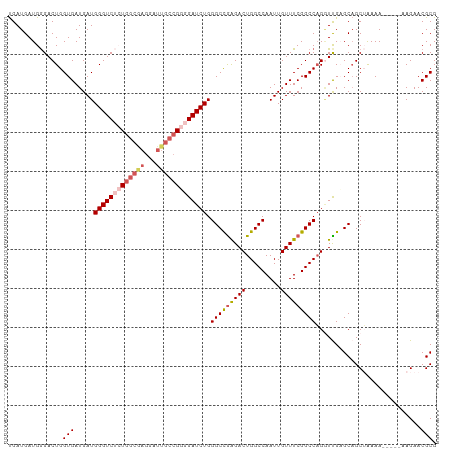
>dm3.chr3R 9766701 95 + 27905053 UGAUGAUGGGACUGGUGAUGAUGGGUCUCUGGC----------GGGGAUCUGGGCCGAGACUGGCCAAUUGUUUCGGCCAGUUUGAGCAGCUAAAA-----AAGAACGCG .......((((((.((....)).))))))..((----------(....(((.(((((((((.........)))))))))((((.....))))....-----.))).))). ( -29.80, z-score = -0.78, R) >droEre2.scaffold_4770 5882966 103 - 17746568 UGAUGAUGAGACUGGUGAUGAUGGGUC--UGGCGAGGAUUGCCGUGGAUCUGGGCCGGGACUGGCCAAUUGUUUCGGCCUGGUCGAGCAGCAAAAA-----AAGAACGCG ........(((((.((....)).))))--).(((....((((.((.((((.((((((..((.........))..))))))))))..)).))))...-----.....))). ( -34.00, z-score = -1.07, R) >droYak2.chr3R 14082012 108 + 28832112 CGAUGAUGGGACUGGUGAUGAUGGGUC--UGGCGAGGAUUGCCGGGGAUCUGGGCUGAGACUGGCCAAUUGUUUCGGCCAGAUCGAGCUGCAAAAACAAAAAAGAACGCG ........(((((.((....)).))))--).(((.((....))...(((((((.(((((((.........))))))))))))))......................))). ( -32.00, z-score = -0.74, R) >droSec1.super_0 8877367 105 - 21120651 UGAUGAUGGGACUGGUGAUGAUGGGUCUCUGGCAAGGAUUGCCGGGGAUCUGGGCCAAGACUGGCCAAUUGUUUCGGCCAGGUUGUGCAGCUGAAA-----AAGAACGCG ...........(((((((..((((((((((((((.....)))))))))))).(((((....))))).))..))...)))))...(((...((....-----.))..))). ( -37.80, z-score = -1.80, R) >droSim1.chr3R 15856811 105 - 27517382 UGAUGAUGGGACUGGUGAUGAUGGGUCUCUGGCGAGGAUUGCCGGGGAUCUGGGCCGAGACUGGCCAAUUGUUUCGGCCAGGUUGUGCAGCUGAAA-----AAGAACGCG ......(.((.((((..((...(((((((((((((...))))))))))))).(((((((((.........)))))))))..))..).))))).)..-----......... ( -38.80, z-score = -2.10, R) >consensus UGAUGAUGGGACUGGUGAUGAUGGGUCUCUGGCGAGGAUUGCCGGGGAUCUGGGCCGAGACUGGCCAAUUGUUUCGGCCAGGUUGAGCAGCUAAAA_____AAGAACGCG ..............(((.....(((((((((((((...))))))))))))).(((((((((.........)))))))))...........................))). (-29.56 = -30.82 + 1.26)

| Location | 9,766,733 – 9,766,828 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 90.90 |
| Shannon entropy | 0.15615 |
| G+C content | 0.51430 |
| Mean single sequence MFE | -26.86 |
| Consensus MFE | -22.23 |
| Energy contribution | -22.55 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
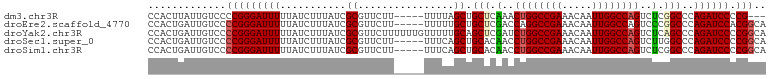
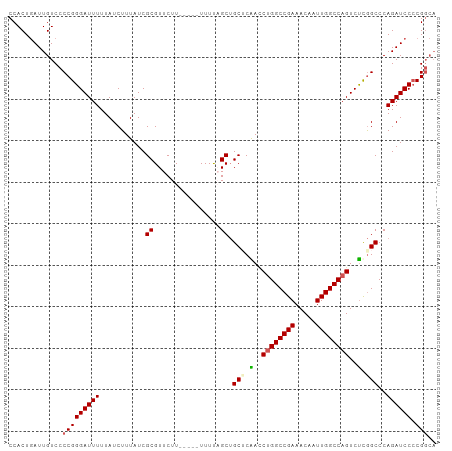
>dm3.chr3R 9766733 95 - 27905053 CCACUUAUUGUCCCCGGGAUUUUUAUCUUUAUCGCGUUCUU-----UUUUAGCUGCUCAAACUGGCCGAAACAAUUGGCCAGUCUCGGCCCAGAUCCCCG--- ...............(((((((...........((......-----.....)).(((.(.(((((((((.....))))))))).).)))..)))))))..--- ( -25.60, z-score = -2.56, R) >droEre2.scaffold_4770 5883003 98 + 17746568 CCACUGAUUGUCCCCGGGAUUUUUAUCUUUAUCGCGUUCUU-----UUUUUGCUGCUCGACCAGGCCGAAACAAUUGGCCAGUCCCGGCCCAGAUCCACGGCA ((...((((....((((((((..........(((.((....-----........)).)))...((((((.....))))))))))))))....))))...)).. ( -27.60, z-score = -1.21, R) >droYak2.chr3R 14082049 103 - 28832112 CCACUGAUUGUCCCCGGGAUUUUUAUCUUUAUCGCGUUCUUUUUUGUUUUUGCAGCUCGAUCUGGCCGAAACAAUUGGCCAGUCUCAGCCCAGAUCCCCGGCA ............((.(((((((...........(((..(......)....))).(((.((.((((((((.....))))))))..)))))..))))))).)).. ( -28.30, z-score = -1.79, R) >droSec1.super_0 8877406 98 + 21120651 CCACUGAUUGUCCCCGGGAUUUUUAUCUUUAUCGCGUUCUU-----UUUCAGCUGCACAACCUGGCCGAAACAAUUGGCCAGUCUUGGCCCAGAUCCCCGGCA ............((.(((((((...........((......-----.....)).((.(((.((((((((.....))))))))..)))))..))))))).)).. ( -27.00, z-score = -1.22, R) >droSim1.chr3R 15856850 98 + 27517382 CCACUGAUUGUCCCCGGGAUUUUUAUCUUUAUCGCGUUCUU-----UUUCAGCUGCACAACCUGGCCGAAACAAUUGGCCAGUCUCGGCCCAGAUCCCCGGCA ............((.(((((((...........((......-----.....)).((.....((((((((.....)))))))).....))..))))))).)).. ( -25.80, z-score = -0.84, R) >consensus CCACUGAUUGUCCCCGGGAUUUUUAUCUUUAUCGCGUUCUU_____UUUUAGCUGCUCAACCUGGCCGAAACAAUUGGCCAGUCUCGGCCCAGAUCCCCGGCA .............(((((((((...........((................)).(((.(..((((((((.....))))))))..).)))..)))))).))).. (-22.23 = -22.55 + 0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:13:48 2011