
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,373,439 – 7,373,540 |
| Length | 101 |
| Max. P | 0.999699 |

| Location | 7,373,439 – 7,373,540 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 82.71 |
| Shannon entropy | 0.30622 |
| G+C content | 0.52305 |
| Mean single sequence MFE | -34.67 |
| Consensus MFE | -22.30 |
| Energy contribution | -22.20 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.969715 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
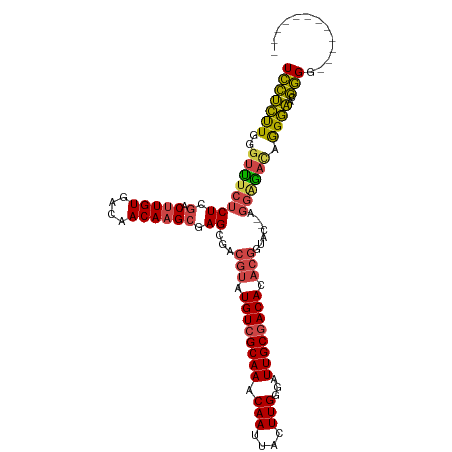
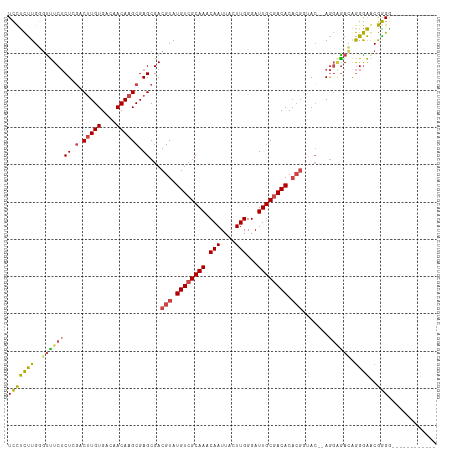
>dm3.chr2L 7373439 101 + 23011544 UCCUCUUGGGUUUCUCUCGACUUGUGACAACAAGCGAGCGACGUAUGUCGCAAACAAUUACUUGGGAUUGCGACACACGGUAC--AGGCGAUGGGGAGCGGGG------------ ((((((((....((.((((.(((((....))))))))).))(((.((((((((.(((....)))...)))))))).)))....--...))).)))))......------------ ( -35.30, z-score = -2.47, R) >droAna3.scaffold_12916 15278480 113 + 16180835 UCUCUCUGUCUCUCGCUGCUCAUGUGACAACAAGCGAGCGACGUAUGUCGCAAACAAUUACUUGGGAUUGCAACACACGAGAC--AACAGACAAGAGACAGGGGCAUGGAGCUUG ...((((((((((.(((((((.((((...........(((((....)))))...(((((......)))))...)))).)))..--..))).).))))))))))((.....))... ( -36.10, z-score = -2.09, R) >droEre2.scaffold_4929 16288492 102 + 26641161 UCCUCUUGGGCUUCUCUGGACUUGUGACAACAAGCGAGCGACGUAUGUCGCAAACAAUUACUUGGGAUUGCGACACACGGGAAGAAGGAGACAGGGAACGGG------------- ..(((((...(((((((.(.(((((....)))))).))...(((.((((((((.(((....)))...)))))))).)))))))).)))))...(....)...------------- ( -32.40, z-score = -2.31, R) >droYak2.chr2L 16794110 96 - 22324452 UCCUCUUGGGUUUCUCUGGACUUGUGACAACAAGCGAGCGACGUAUGUCGCAAACAAUUACUUGGGAUUGCGACAC-----AC--AGGGAACAGGGAACGGGA------------ ...(((((...(((((((..((((((.......(((.....))).((((((((.(((....)))...)))))))))-----))--)))...))))))))))))------------ ( -31.50, z-score = -2.66, R) >droSec1.super_3 2888483 101 + 7220098 UUCUCUUGGGUUUCUCUCGACUUGUGACAACAAGCGAGCGACGUAUGUCGCAAACAAUUACUUGGGAUUGCGACACACGGUAC--AGGAGAUGGGGAGCGAGG------------ .(((((((....((.((((.(((((....))))))))).))(((.((((((((.(((....)))...)))))))).)))...)--))))))............------------ ( -37.00, z-score = -3.68, R) >droSim1.chr2L 7161573 101 + 22036055 UCCUCUUGGGUUUCUCUCGACUUGUGACAACAAGCGAGCGACGUAUGUCGCAAACAAUUACUUGGGAUUGCGACACACGGUAC--AGGAGAUGGGGAGCGGGG------------ ..((((((....((.((((.(((((....))))))))).))(((.((((((((.(((....)))...)))))))).)))...)--))))).............------------ ( -35.70, z-score = -2.79, R) >consensus UCCUCUUGGGUUUCUCUCGACUUGUGACAACAAGCGAGCGACGUAUGUCGCAAACAAUUACUUGGGAUUGCGACACACGGUAC__AGGAGACAGGGAACGGGG____________ (((((((..((((((((.(.(((((....)))))).))...(((.((((((((.(((....)))...)))))))).))).......)))))).))))..)))............. (-22.30 = -22.20 + -0.10)

| Location | 7,373,439 – 7,373,540 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 82.71 |
| Shannon entropy | 0.30622 |
| G+C content | 0.52305 |
| Mean single sequence MFE | -35.90 |
| Consensus MFE | -25.37 |
| Energy contribution | -25.02 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.21 |
| Mean z-score | -5.07 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.21 |
| SVM RNA-class probability | 0.999699 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
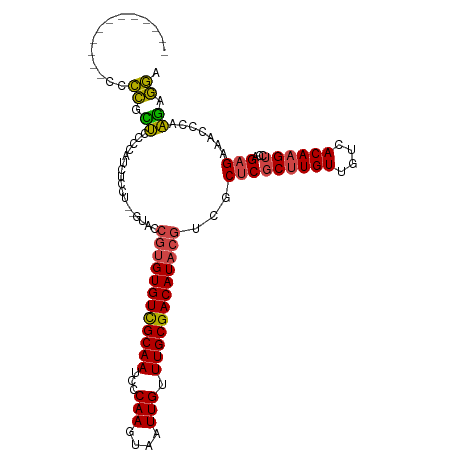
>dm3.chr2L 7373439 101 - 23011544 ------------CCCCGCUCCCCAUCGCCU--GUACCGUGUGUCGCAAUCCCAAGUAAUUGUUUGCGACAUACGUCGCUCGCUUGUUGUCACAAGUCGAGAGAAACCCAAGAGGA ------------...............(((--.(..((((((((((((...(((....))).))))))))))))((.(((((((((....))))).)))).))......).))). ( -35.00, z-score = -4.77, R) >droAna3.scaffold_12916 15278480 113 - 16180835 CAAGCUCCAUGCCCCUGUCUCUUGUCUGUU--GUCUCGUGUGUUGCAAUCCCAAGUAAUUGUUUGCGACAUACGUCGCUCGCUUGUUGUCACAUGAGCAGCGAGAGACAGAGAGA ...((.....))..(((((((((((.....--....((((((((((((...(((....))).))))))))))))..(((((..((....))..))))).)))))))))))..... ( -43.60, z-score = -4.30, R) >droEre2.scaffold_4929 16288492 102 - 26641161 -------------CCCGUUCCCUGUCUCCUUCUUCCCGUGUGUCGCAAUCCCAAGUAAUUGUUUGCGACAUACGUCGCUCGCUUGUUGUCACAAGUCCAGAGAAGCCCAAGAGGA -------------.....(((((.....((((((..((((((((((((...(((....))).))))))))))))......((((((....))))))...))))))....)).))) ( -33.50, z-score = -5.07, R) >droYak2.chr2L 16794110 96 + 22324452 ------------UCCCGUUCCCUGUUCCCU--GU-----GUGUCGCAAUCCCAAGUAAUUGUUUGCGACAUACGUCGCUCGCUUGUUGUCACAAGUCCAGAGAAACCCAAGAGGA ------------........(((.((....--((-----(((((((((...(((....))).)))))))))))((..(((((((((....))))))...)))..))..)).))). ( -30.60, z-score = -4.53, R) >droSec1.super_3 2888483 101 - 7220098 ------------CCUCGCUCCCCAUCUCCU--GUACCGUGUGUCGCAAUCCCAAGUAAUUGUUUGCGACAUACGUCGCUCGCUUGUUGUCACAAGUCGAGAGAAACCCAAGAGAA ------------............((((.(--(...((((((((((((...(((....))).))))))))))))((.(((((((((....))))).)))).))....)).)))). ( -36.80, z-score = -6.26, R) >droSim1.chr2L 7161573 101 - 22036055 ------------CCCCGCUCCCCAUCUCCU--GUACCGUGUGUCGCAAUCCCAAGUAAUUGUUUGCGACAUACGUCGCUCGCUUGUUGUCACAAGUCGAGAGAAACCCAAGAGGA ------------............((((.(--(...((((((((((((...(((....))).))))))))))))((.(((((((((....))))).)))).))....)).)))). ( -35.90, z-score = -5.47, R) >consensus ____________CCCCGCUCCCCAUCUCCU__GUACCGUGUGUCGCAAUCCCAAGUAAUUGUUUGCGACAUACGUCGCUCGCUUGUUGUCACAAGUCCAGAGAAACCCAAGAGGA ..............((.((.................((((((((((((...(((....))).))))))))))))...(((((((((....))))))...))).......)).)). (-25.37 = -25.02 + -0.36)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:23:32 2011