
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,759,108 – 9,759,199 |
| Length | 91 |
| Max. P | 0.994414 |

| Location | 9,759,108 – 9,759,199 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 66.95 |
| Shannon entropy | 0.60233 |
| G+C content | 0.50031 |
| Mean single sequence MFE | -22.61 |
| Consensus MFE | -13.39 |
| Energy contribution | -13.01 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.70 |
| SVM RNA-class probability | 0.994414 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
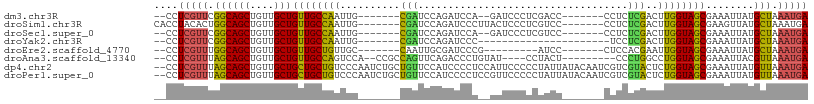
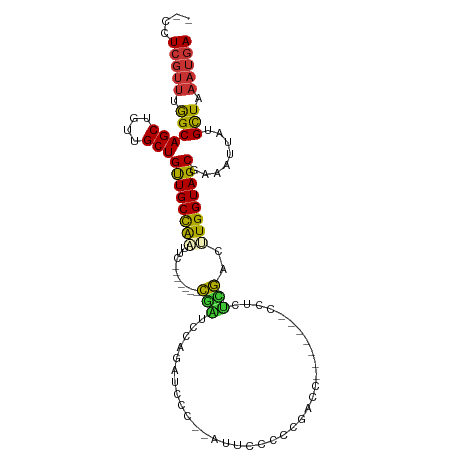
>dm3.chr3R 9759108 91 + 27905053 --CCUCGUUCGGCAGCUGUUGCUGUUGCCAAUUG-------CGAUCCAGAUCCA--GAUCCCUCGACC-------CCUCUCGACUUGGUAGCGAAAUUAUGCUAAAUGA --..(((((..(((...(((..(((((((((...-------.((((........--))))..((((..-------....)))).))))))))).)))..)))..))))) ( -25.00, z-score = -1.89, R) >droSim1.chr3R 15849262 95 - 27517382 CACCUACACUGGCAGCUGUUGCUGUUGCCAAUUG-------CGAUCCAGAUCCCUUACUCCCUCGUCC-------CCUCUCGACUUGGUAGCGAAGUUAUGCUAAAUGA .........(((((((.......)))))))...(-------((((....)))..........((((..-------((.........))..))))......))....... ( -18.60, z-score = 0.03, R) >droSec1.super_0 8869862 91 - 21120651 --CCUCGUUCGGCAGCUGUUGCUGUUGCCAAUUG-------CGAUCCAGAUCCA--GAUCCCUCGUCC-------CCUCUCGACUUGGUAGCGAAAUUAUGCUAAAUGA --..(((((..(((...(((..(((((((((...-------.((((........--))))....(((.-------......)))))))))))).)))..)))..))))) ( -24.00, z-score = -1.61, R) >droYak2.chr3R 14074607 78 + 28832112 --CCUCGUUCGGCAGCUGUUGCUGUUGCCAAUUG-------CGAUCCAGAUCCC----------------------UCCUCGACUUGGUAGCGAAAUUAUGCUAAAUGA --..(((((..(((...(((..(((((((((...-------(((...((....)----------------------)..)))..))))))))).)))..)))..))))) ( -22.10, z-score = -1.92, R) >droEre2.scaffold_4770 5875589 84 - 17746568 --CCUCGUUUGGCAGCUGUUGCUGUUGCUGUUGC-------CAAUUGCGAUCCCG---------AUCC-------CUCCACGAAUUGGUAGCGAAAUUAUGCUAAAUGA --..((((((((((((....))).....((((((-------(((((.((......---------....-------.....))))))))))))).......))))))))) ( -24.46, z-score = -2.45, R) >droAna3.scaffold_13340 1114485 92 - 23697760 --CCUCGUUUAGCAGCUGUUGCUGUUGCCAGUCCA--CCGCCAGUUCAGACCCUGUAU----CCUACU---------CCCUGGCCUGGUAGCGAAAUUACGUUAAAUGA --..((((((((((((....)))((((((((....--..(((((..............----......---------..)))))))))))))........))))))))) ( -26.75, z-score = -2.57, R) >dp4.chr2 17147596 107 - 30794189 --CCUCGUUUAGCAGCUGUUGCUGCUGCUGUCCCAAUCUGCUGUUCCAUCCCCUCCAUUCCCCCUAUUAUACAAUCGUCGUACUCUGGUAGCGAAAUUAUGUUAAAUGA --..((((((((((....((((((((((...........))............................(((.......)))....)))))))).....)))))))))) ( -20.00, z-score = -2.16, R) >droPer1.super_0 6800689 107 + 11822988 --CCUCGUUUAGCAGCUGUUGCUGCUGCUGUCCCAAUCUGCUGUUCCAUCCCCUCCGUUCCCCCUAUUAUACAAUCGUCGUACUCUGGUAGCGAAAUUAUGUUAAAUGA --..((((((((((....((((((((((...........))............................(((.......)))....)))))))).....)))))))))) ( -20.00, z-score = -2.06, R) >consensus __CCUCGUUUGGCAGCUGUUGCUGUUGCCAAUUC_______CGAUCCAGAUCCC__AUUCCCCCGACC_______CCUCUCGACUUGGUAGCGAAAUUAUGCUAAAUGA ....(((((.((((((....)))((((((((.....................................................))))))))........))).))))) (-13.39 = -13.01 + -0.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:13:44 2011