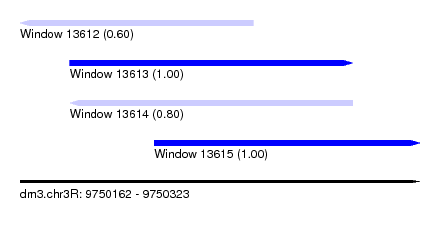
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,750,162 – 9,750,323 |
| Length | 161 |
| Max. P | 0.998111 |

| Location | 9,750,162 – 9,750,256 |
|---|---|
| Length | 94 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 63.22 |
| Shannon entropy | 0.74609 |
| G+C content | 0.42813 |
| Mean single sequence MFE | -13.40 |
| Consensus MFE | -7.28 |
| Energy contribution | -6.73 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.31 |
| Mean z-score | -0.11 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.595928 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9750162 94 - 27905053 UAAAUGACAAACGAUUUUGUGAAAUUAUUAUUGCCUUUGUCAUCAUCGCUUGA--CAGUC---AGACCCACU-CACCCCAG---AAAUGCGUCCUUCCUCAGC-------------- ....((((...((((..((......))..))))....(((((........)))--)))))---)(((.((.(-(......)---)..)).)))..........-------------- ( -11.90, z-score = 0.29, R) >droSim1.chr3R 15839979 97 + 27517382 UAAAUGACAAACGAUUUUGUGAAAUUAUUAUUGCCUUUGUCAUCAUCGCUCGA--CAGUCAGCAGACCCACU-CACCCCAG---AAAUUCGUCCUUCCUCAGC-------------- ..........((((.(((.((..........(((..(((((..........))--)))...)))((.....)-)....)).---))).))))...........-------------- ( -11.70, z-score = 0.39, R) >droSec1.super_0 8861077 97 + 21120651 UAAAUGACAAACGAUUUUGUGAAAUUAUUAUUGCCUUUGUCAUCAUCGCUCGA--CAGUCAGCAGACCCACU-CACCCCAG---AAAUUCGUCCUUCCUCAGC-------------- ..........((((.(((.((..........(((..(((((..........))--)))...)))((.....)-)....)).---))).))))...........-------------- ( -11.70, z-score = 0.39, R) >droYak2.chr3R 14065569 96 - 28832112 UAAAUGACAAACGAUUUUGUGAAAUUAUUAUUGCCUUUGUCAUCAUCGCUCGA--CAGUC---AGAUCCACU-CACCCCAU-CCUCACUCGUCCUUUCUCUCC-------------- ...((((((((((((..((......))..))))..))))))))........((--((((.---((.......-........-.)).))).)))..........-------------- ( -11.19, z-score = -0.52, R) >droEre2.scaffold_4770 5866411 94 + 17746568 UAAAUGACAAACGAUUUUGUGAAAUUAUUAUUGCCUUUGUCAUCAUCGCUCGA--CAGUC---AGAUCCACU-CACCCCAG---ACACUCGUCCUUUCUCUCC-------------- ...((((((((((((..((......))..))))..))))))))....(..(((--..(((---.........-.......)---))..)))..).........-------------- ( -10.89, z-score = 0.28, R) >droAna3.scaffold_13340 1104656 106 + 23697760 UAAAUGACAAACGAUUUUGUGAAAUUAUUAUUGCCAUUGUCAUCAUCGCUUGG--CUGCC---ACUUUCACUCCAUCCCGAACUUCAUCAGCCUCGAGUCCUCCCAUCUCC------ ...(((((((.((((..((......))..))))...)))))))..(((...((--(((..---...(((..........)))......))))).)))..............------ ( -15.80, z-score = -0.77, R) >droPer1.super_0 6791869 115 - 11822988 UAAAUGACAAACGAUUUUGUGAAAUUAUUAUUGCCAUUGUCAUCAUCGCUUGGCCUAGCUG--CCUGACGACCCACUCCACAACUUCAUCAACGCCAUCCUUUGGUCUUCCUUCCCC ...(((((((.((((..((......))..))))...)))))))..(((...(((......)--))...)))......................((((.....))))........... ( -16.20, z-score = -0.15, R) >dp4.chr2 17138849 115 + 30794189 UAAAUGACAAACGAUUUUGUGAAAUUAUUAUUGCCAUUGUCAUCAUCGCUUGGCCUAGCUG--CCUGACGACCCACUCCACAACUUCAUCAACGCCAUCCUUUGGUCUUCCUUCCCC ...(((((((.((((..((......))..))))...)))))))..(((...(((......)--))...)))......................((((.....))))........... ( -16.20, z-score = -0.15, R) >droWil1.scaffold_181108 2824951 80 + 4707319 UAAAUGACAAACGAUUUUGUGAAAUUAUUAUUGCCAUUGUCAUCAUCUGAUUCUCCUUCCA-UUCUCCAUCUUUAUUCUCA------------------------------------ ...(((((((.((((..((......))..))))...)))))))..................-...................------------------------------------ ( -7.00, z-score = -0.20, R) >droVir3.scaffold_13047 500942 83 + 19223366 UAAAUGACAAACGAUUUUGCAAAAUUAUUAUUGCCAUUGUCAUCGUCGCUUGG-UUGGCUGACUCUGGCCCCUGACUCU--CCAGC------------------------------- ...(((((((.((((..((......))..))))...))))))).((((...((-(..(......)..)))..))))...--.....------------------------------- ( -15.00, z-score = 0.10, R) >droMoj3.scaffold_6540 19693235 80 - 34148556 UAAAUGACAAACGAUUUUGCAAAAUUAUUAUUGCCAUUGUCAUCGUCGUUCGG-UUGGCUGGCUGCGUCU-CUGACCCCAAA----------------------------------- ..((((((..(((((...((((........)))).)))))....)))))).((-(..(..(((...))).-)..))).....----------------------------------- ( -15.10, z-score = 0.08, R) >droGri2.scaffold_14830 2415705 93 + 6267026 UAAAUGACAAACGAUUUUGCGAAAUUAUUAUUGCCAUUGUCAUCGUCGCUUGG-UUGGCUGAUUCCGUCUCCCGACUCCUGUCACUUGUCCCCC----------------------- .....(((((..(((..(((((........))).))..)))...((.((..((-((((..(((...)))..))))))...)).)))))))....----------------------- ( -18.10, z-score = -1.10, R) >consensus UAAAUGACAAACGAUUUUGUGAAAUUAUUAUUGCCAUUGUCAUCAUCGCUUGG__CAGCCG__ACAGCCACU_CACCCCAG___ACAUUCGUCCUU_CUC__C______________ ...(((((((.((((..((......))..))))...)))))))....(((......))).......................................................... ( -7.28 = -6.73 + -0.55)

| Location | 9,750,182 – 9,750,296 |
|---|---|
| Length | 114 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 84.80 |
| Shannon entropy | 0.34033 |
| G+C content | 0.44602 |
| Mean single sequence MFE | -36.26 |
| Consensus MFE | -30.64 |
| Energy contribution | -30.72 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.26 |
| SVM RNA-class probability | 0.998111 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9750182 114 + 27905053 UGGGGUG-AGUGGGUCUG----ACUGUCAAGCGAUGAUGACAAAGGCAAUAAUAAUUUCACAAAAUCGUUUGUCAUUUACGCAUGGCAAACGGUUUUGAAAGUAAACAGCCGGGUCGUC ...(((.-((.....)).----)))(((....)))((((((...(((.............((((((((((((((((......))))))))))))))))...(....).)))..)))))) ( -35.00, z-score = -2.92, R) >droSim1.chr3R 15839999 117 - 27517382 UGGGGUG-AGUGGGUCUGC-UGACUGUCGAGCGAUGAUGACAAAGGCAAUAAUAAUUUCACAAAAUCGUUUGUCAUUUACGCAUGGCAAACGGUUUUGAAAGUAAACAGCCGGGUCGUC ....((.-.(..((((...-.))))..)..))...((((((...(((.............((((((((((((((((......))))))))))))))))...(....).)))..)))))) ( -37.40, z-score = -2.77, R) >droSec1.super_0 8861097 117 - 21120651 UGGGGUG-AGUGGGUCUGC-UGACUGUCGAGCGAUGAUGACAAAGGCAAUAAUAAUUUCACAAAAUCGUUUGUCAUUUACGCAUGGCAAACGGUUUUGAAAGUAAACAGCCGGGUCGUC ....((.-.(..((((...-.))))..)..))...((((((...(((.............((((((((((((((((......))))))))))))))))...(....).)))..)))))) ( -37.40, z-score = -2.77, R) >droYak2.chr3R 14065591 114 + 28832112 UGGGGUG-AGUGGAUCUG----ACUGUCGAGCGAUGAUGACAAAGGCAAUAAUAAUUUCACAAAAUCGUUUGUCAUUUACGCAUGGCAAACGGUUUUGAAAGUAAACAGCCGGGUCGUC ...(((.-((.....)).----)))(((....)))((((((...(((.............((((((((((((((((......))))))))))))))))...(....).)))..)))))) ( -35.50, z-score = -2.87, R) >droEre2.scaffold_4770 5866431 114 - 17746568 UGGGGUG-AGUGGAUCUG----ACUGUCGAGCGAUGAUGACAAAGGCAAUAAUAAUUUCACAAAAUCGUUUGUCAUUUACGCAUGGCAAACGGUUUUGAAAGUAAACAGCCGGGUCGUC ...(((.-((.....)).----)))(((....)))((((((...(((.............((((((((((((((((......))))))))))))))))...(....).)))..)))))) ( -35.50, z-score = -2.87, R) >droAna3.scaffold_13340 1104688 114 - 23697760 -GGGAUGGAGUGAAAGUG----GCAGCCAAGCGAUGAUGACAAUGGCAAUAAUAAUUUCACAAAAUCGUUUGUCAUUUACGCAUGGCAAACGGUUUUGAAAGUAAACAGCCGGGUCGUC -..............((.----((......)).))((((((..((((.............((((((((((((((((......))))))))))))))))...(....).)))).)))))) ( -33.00, z-score = -2.34, R) >droPer1.super_0 6791906 118 + 11822988 UGGAGUGGGUCGUCAGGCA-GCUAGGCCAAGCGAUGAUGACAAUGGCAAUAAUAAUUUCACAAAAUCGUUUGUCAUUUACGCAUGGCAAACGGUUUUGAAAGUAAACAGCCGGGUCGUC ........(((((..(((.-.....)))..)))))((((((..((((.............((((((((((((((((......))))))))))))))))...(....).)))).)))))) ( -41.00, z-score = -4.23, R) >dp4.chr2 17138886 118 - 30794189 UGGAGUGGGUCGUCAGGCA-GCUAGGCCAAGCGAUGAUGACAAUGGCAAUAAUAAUUUCACAAAAUCGUUUGUCAUUUACGCAUGGCAAACGGUUUUGAAAGUAAACAGCCGGGUCGUC ........(((((..(((.-.....)))..)))))((((((..((((.............((((((((((((((((......))))))))))))))))...(....).)))).)))))) ( -41.00, z-score = -4.23, R) >droWil1.scaffold_181108 2824970 101 - 4707319 --------------UGGAA-GGAGAAUCA---GAUGAUGACAAUGGCAAUAAUAAUUUCACAAAAUCGUUUGUCAUUUACGCAUGGCAAACGGUUUUGAAAGUAAACAGCCGGGUCUUC --------------.....-(((((.(((---.....)))...((((.............((((((((((((((((......))))))))))))))))...(....).))))..))))) ( -27.60, z-score = -3.11, R) >droVir3.scaffold_13047 500946 119 - 19223366 GAGAGUCAGGGGCCAGAGUCAGCCAACCAAGCGACGAUGACAAUGGCAAUAAUAAUUUUGCAAAAUCGUUUGUCAUUUACGCAUGGCAAACGGUUUUGAAAGUAAACAGCCGGGUCGUC ....(((.(((((........)))..))....)))((((((..((((.......(((((.((((((((((((((((......))))))))))))))))))))).....)))).)))))) ( -37.60, z-score = -3.25, R) >droMoj3.scaffold_6540 19693237 118 + 34148556 UGGGGUCAG-AGACGCAGCCAGCCAACCGAACGACGAUGACAAUGGCAAUAAUAAUUUUGCAAAAUCGUUUGUCAUUUACGCAUGGCAAACGGUUUUGAAAGUAAACAGCCGGGUCGUC ..(((((..-.)))((.....))...)).......((((((..((((.......(((((.((((((((((((((((......))))))))))))))))))))).....)))).)))))) ( -35.00, z-score = -1.90, R) >droGri2.scaffold_14830 2415719 119 - 6267026 AGGAGUCGGGAGACGGAAUCAGCCAACCAAGCGACGAUGACAAUGGCAAUAAUAAUUUCGCAAAAUCGUUUGUCAUUUACGCAUGGCAAACGGUUUUGAAAGUAAACAGCCGGGUCGUC ....(((((....)((......)).......))))((((((..((((.............((((((((((((((((......))))))))))))))))...(....).)))).)))))) ( -39.10, z-score = -3.67, R) >consensus UGGGGUG_AGUGGCUCUG___GACAGCCAAGCGAUGAUGACAAUGGCAAUAAUAAUUUCACAAAAUCGUUUGUCAUUUACGCAUGGCAAACGGUUUUGAAAGUAAACAGCCGGGUCGUC ...................................((((((...(((.............((((((((((((((((......))))))))))))))))...(....).)))..)))))) (-30.64 = -30.72 + 0.08)

| Location | 9,750,182 – 9,750,296 |
|---|---|
| Length | 114 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 84.80 |
| Shannon entropy | 0.34033 |
| G+C content | 0.44602 |
| Mean single sequence MFE | -24.71 |
| Consensus MFE | -19.47 |
| Energy contribution | -19.46 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.801280 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
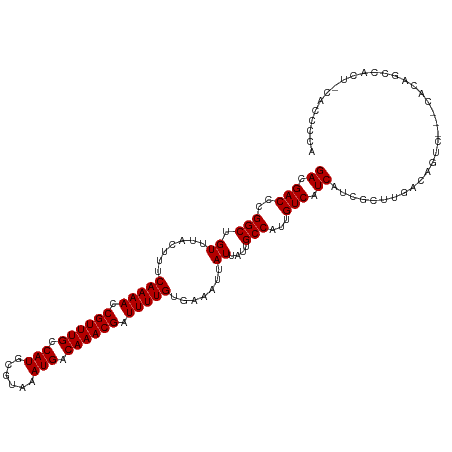
>dm3.chr3R 9750182 114 - 27905053 GACGACCCGGCUGUUUACUUUCAAAACCGUUUGCCAUGCGUAAAUGACAAACGAUUUUGUGAAAUUAUUAUUGCCUUUGUCAUCAUCGCUUGACAGU----CAGACCCACU-CACCCCA (((.....(((....((.((((((((.((((((.(((......))).)))))).)))..))))).)).....)))..(((((........)))))))----).........-....... ( -24.50, z-score = -2.08, R) >droSim1.chr3R 15839999 117 + 27517382 GACGACCCGGCUGUUUACUUUCAAAACCGUUUGCCAUGCGUAAAUGACAAACGAUUUUGUGAAAUUAUUAUUGCCUUUGUCAUCAUCGCUCGACAGUCA-GCAGACCCACU-CACCCCA ...((...(((((((.(((.((......(....)...(((...((((((((((((..((......))..))))..))))))))...)))..)).))).)-)))).))...)-)...... ( -23.40, z-score = -1.18, R) >droSec1.super_0 8861097 117 + 21120651 GACGACCCGGCUGUUUACUUUCAAAACCGUUUGCCAUGCGUAAAUGACAAACGAUUUUGUGAAAUUAUUAUUGCCUUUGUCAUCAUCGCUCGACAGUCA-GCAGACCCACU-CACCCCA ...((...(((((((.(((.((......(....)...(((...((((((((((((..((......))..))))..))))))))...)))..)).))).)-)))).))...)-)...... ( -23.40, z-score = -1.18, R) >droYak2.chr3R 14065591 114 - 28832112 GACGACCCGGCUGUUUACUUUCAAAACCGUUUGCCAUGCGUAAAUGACAAACGAUUUUGUGAAAUUAUUAUUGCCUUUGUCAUCAUCGCUCGACAGU----CAGAUCCACU-CACCCCA (((.....(((....((.((((((((.((((((.(((......))).)))))).)))..))))).)).....)))..((((..........))))))----).........-....... ( -21.90, z-score = -1.03, R) >droEre2.scaffold_4770 5866431 114 + 17746568 GACGACCCGGCUGUUUACUUUCAAAACCGUUUGCCAUGCGUAAAUGACAAACGAUUUUGUGAAAUUAUUAUUGCCUUUGUCAUCAUCGCUCGACAGU----CAGAUCCACU-CACCCCA (((.....(((....((.((((((((.((((((.(((......))).)))))).)))..))))).)).....)))..((((..........))))))----).........-....... ( -21.90, z-score = -1.03, R) >droAna3.scaffold_13340 1104688 114 + 23697760 GACGACCCGGCUGUUUACUUUCAAAACCGUUUGCCAUGCGUAAAUGACAAACGAUUUUGUGAAAUUAUUAUUGCCAUUGUCAUCAUCGCUUGGCUGC----CACUUUCACUCCAUCCC- ........(((.((((.......)))).....((((.(((...(((((((.((((..((......))..))))...)))))))...))).)))).))----)................- ( -24.50, z-score = -1.86, R) >droPer1.super_0 6791906 118 - 11822988 GACGACCCGGCUGUUUACUUUCAAAACCGUUUGCCAUGCGUAAAUGACAAACGAUUUUGUGAAAUUAUUAUUGCCAUUGUCAUCAUCGCUUGGCCUAGC-UGCCUGACGACCCACUCCA (((((...(((....((.((((((((.((((((.(((......))).)))))).)))..))))).)).....))).)))))....(((...(((.....-.)))...)))......... ( -26.10, z-score = -1.60, R) >dp4.chr2 17138886 118 + 30794189 GACGACCCGGCUGUUUACUUUCAAAACCGUUUGCCAUGCGUAAAUGACAAACGAUUUUGUGAAAUUAUUAUUGCCAUUGUCAUCAUCGCUUGGCCUAGC-UGCCUGACGACCCACUCCA (((((...(((....((.((((((((.((((((.(((......))).)))))).)))..))))).)).....))).)))))....(((...(((.....-.)))...)))......... ( -26.10, z-score = -1.60, R) >droWil1.scaffold_181108 2824970 101 + 4707319 GAAGACCCGGCUGUUUACUUUCAAAACCGUUUGCCAUGCGUAAAUGACAAACGAUUUUGUGAAAUUAUUAUUGCCAUUGUCAUCAUC---UGAUUCUCC-UUCCA-------------- ((((....(((....((.((((((((.((((((.(((......))).)))))).)))..))))).)).....)))...((((.....---))))....)-)))..-------------- ( -20.00, z-score = -1.90, R) >droVir3.scaffold_13047 500946 119 + 19223366 GACGACCCGGCUGUUUACUUUCAAAACCGUUUGCCAUGCGUAAAUGACAAACGAUUUUGCAAAAUUAUUAUUGCCAUUGUCAUCGUCGCUUGGUUGGCUGACUCUGGCCCCUGACUCUC (((((...(((.((....((((((((.((((((.(((......))).)))))).))))).)))......)).))).)))))...((((...(((..(......)..)))..)))).... ( -28.70, z-score = -1.56, R) >droMoj3.scaffold_6540 19693237 118 - 34148556 GACGACCCGGCUGUUUACUUUCAAAACCGUUUGCCAUGCGUAAAUGACAAACGAUUUUGCAAAAUUAUUAUUGCCAUUGUCAUCGUCGUUCGGUUGGCUGGCUGCGUCU-CUGACCCCA ((.(((.(((((......((((((((.((((((.(((......))).)))))).))))).))).........((((.....((((.....)))))))).))))).))))-)........ ( -28.80, z-score = -0.60, R) >droGri2.scaffold_14830 2415719 119 + 6267026 GACGACCCGGCUGUUUACUUUCAAAACCGUUUGCCAUGCGUAAAUGACAAACGAUUUUGCGAAAUUAUUAUUGCCAUUGUCAUCGUCGCUUGGUUGGCUGAUUCCGUCUCCCGACUCCU (((((...(((....((.((((((((.((((((.(((......))).)))))).))))..)))).)).....))).......)))))....((((((..(((...)))..))))))... ( -27.20, z-score = -0.97, R) >consensus GACGACCCGGCUGUUUACUUUCAAAACCGUUUGCCAUGCGUAAAUGACAAACGAUUUUGUGAAAUUAUUAUUGCCAUUGUCAUCAUCGCUUGACAGUC___CACAGCCACU_CACCCCA ((.(((..(((.((.......(((((.((((((.(((......))).)))))).))))).......))....)))...))).))................................... (-19.47 = -19.46 + -0.00)
| Location | 9,750,216 – 9,750,323 |
|---|---|
| Length | 107 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 86.43 |
| Shannon entropy | 0.30103 |
| G+C content | 0.43553 |
| Mean single sequence MFE | -33.51 |
| Consensus MFE | -25.82 |
| Energy contribution | -25.38 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.30 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.18 |
| SVM RNA-class probability | 0.997783 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9750216 107 + 27905053 ACAAAGGCAAUAAUAAUUUCACAAAAUCGUUUGUCAUUUACGCAUGGCAAACGGUUUUGAAAGUAAACAGCCGGGUCGUCAUGGGUCUCUGGUAUUCCACGAGAUGG-------- .....(((.............((((((((((((((((......))))))))))))))))...(....).)))............((((((((....))).)))))..-------- ( -32.30, z-score = -3.61, R) >droSim1.chr3R 15840036 107 - 27517382 ACAAAGGCAAUAAUAAUUUCACAAAAUCGUUUGUCAUUUACGCAUGGCAAACGGUUUUGAAAGUAAACAGCCGGGUCGUCAUGGGUCUCUGGUAUUCCACGAGAUGG-------- .....(((.............((((((((((((((((......))))))))))))))))...(....).)))............((((((((....))).)))))..-------- ( -32.30, z-score = -3.61, R) >droSec1.super_0 8861134 107 - 21120651 ACAAAGGCAAUAAUAAUUUCACAAAAUCGUUUGUCAUUUACGCAUGGCAAACGGUUUUGAAAGUAAACAGCCGGGUCGUCAUGGGUCUCUGGUAUUCCACGAGAUGG-------- .....(((.............((((((((((((((((......))))))))))))))))...(....).)))............((((((((....))).)))))..-------- ( -32.30, z-score = -3.61, R) >droYak2.chr3R 14065625 107 + 28832112 ACAAAGGCAAUAAUAAUUUCACAAAAUCGUUUGUCAUUUACGCAUGGCAAACGGUUUUGAAAGUAAACAGCCGGGUCGUCAUGGGUCUCUGGAAUUCCACGAGAUGG-------- .....(((.............((((((((((((((((......))))))))))))))))...(....).)))............((((((((....))).)))))..-------- ( -32.90, z-score = -3.72, R) >droEre2.scaffold_4770 5866465 107 - 17746568 ACAAAGGCAAUAAUAAUUUCACAAAAUCGUUUGUCAUUUACGCAUGGCAAACGGUUUUGAAAGUAAACAGCCGGGUCGUCAUGGGUCUCUGGUAUUCCACGAGAUGG-------- .....(((.............((((((((((((((((......))))))))))))))))...(....).)))............((((((((....))).)))))..-------- ( -32.30, z-score = -3.61, R) >droAna3.scaffold_13340 1104722 109 - 23697760 ACAAUGGCAAUAAUAAUUUCACAAAAUCGUUUGUCAUUUACGCAUGGCAAACGGUUUUGAAAGUAAACAGCCGGGUCGUCACGGGUUCCCGACAUUUAACGAGAUGUGG------ ....((((.............((((((((((((((((......))))))))))))))))...(....).))))........(((....)))((((((....))))))..------ ( -31.40, z-score = -2.47, R) >droPer1.super_0 6791944 107 + 11822988 ACAAUGGCAAUAAUAAUUUCACAAAAUCGUUUGUCAUUUACGCAUGGCAAACGGUUUUGAAAGUAAACAGCCGGGUCGUCAUGGGUCCCGGACACGCCGCGAGAUGG-------- .....(((.............((((((((((((((((......))))))))))))))))..........((((((((......)).))))).)..))).........-------- ( -35.70, z-score = -2.98, R) >dp4.chr2 17138924 107 - 30794189 ACAAUGGCAAUAAUAAUUUCACAAAAUCGUUUGUCAUUUACGCAUGGCAAACGGUUUUGAAAGUAAACAGCCGGGUCGUCAUGGGUCCCGGACACGCCGCGAGAUGG-------- .....(((.............((((((((((((((((......))))))))))))))))..........((((((((......)).))))).)..))).........-------- ( -35.70, z-score = -2.98, R) >droWil1.scaffold_181108 2824991 114 - 4707319 ACAAUGGCAAUAAUAAUUUCACAAAAUCGUUUGUCAUUUACGCAUGGCAAACGGUUUUGAAAGUAAACAGCCGGGUCUUCAUGGGUCUAUGG-ACAGAAAUACGUGCCUAGGGGG .(...((((......(((((.((((((((((((((((......))))))))))))))))...((...((...((..((....))..)).)).-)).)))))...))))...)... ( -34.00, z-score = -3.38, R) >droVir3.scaffold_13047 500985 102 - 19223366 ACAAUGGCAAUAAUAAUUUUGCAAAAUCGUUUGUCAUUUACGCAUGGCAAACGGUUUUGAAAGUAAACAGCCGGGUCGUCAUGGACCCCAGUGCCAGCAUGC------------- ....(((((......(((((.((((((((((((((((......)))))))))))))))))))))........(((((......)))))...)))))......------------- ( -36.90, z-score = -3.93, R) >droMoj3.scaffold_6540 19693275 105 + 34148556 ACAAUGGCAAUAAUAAUUUUGCAAAAUCGUUUGUCAUUUACGCAUGGCAAACGGUUUUGAAAGUAAACAGCCGGGUCGUCAUCAGCCAUGAUGGGGAGAGGAUGC---------- ....((((.......(((((.((((((((((((((((......))))))))))))))))))))).....))))..((.((((((....)))))).))........---------- ( -33.40, z-score = -3.37, R) >droGri2.scaffold_14830 2415758 105 - 6267026 ACAAUGGCAAUAAUAAUUUCGCAAAAUCGUUUGUCAUUUACGCAUGGCAAACGGUUUUGAAAGUAAACAGCCGGGUCGUCAUGGACCCCAGACUCCGGGGGCAGG---------- ......((.............((((((((((((((((......))))))))))))))))...........(((((..(((.(((...)))))))))))..))...---------- ( -32.90, z-score = -2.30, R) >consensus ACAAUGGCAAUAAUAAUUUCACAAAAUCGUUUGUCAUUUACGCAUGGCAAACGGUUUUGAAAGUAAACAGCCGGGUCGUCAUGGGUCUCUGGCAUUCCACGAGAUGG________ .....(((.............((((((((((((((((......))))))))))))))))...(....).)))(((((......)))))........................... (-25.82 = -25.38 + -0.44)

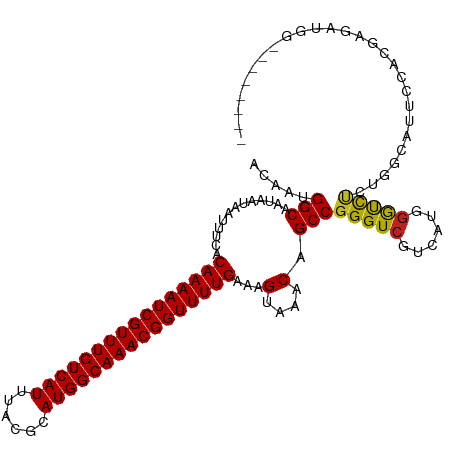
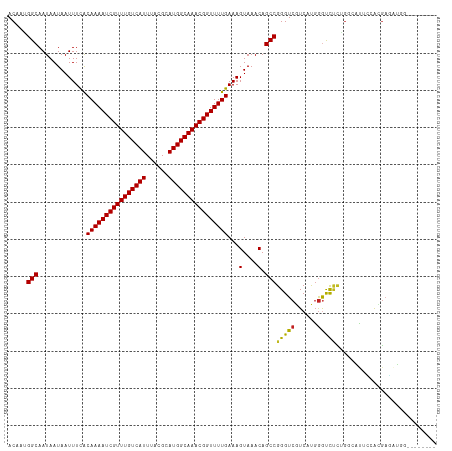
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:13:42 2011