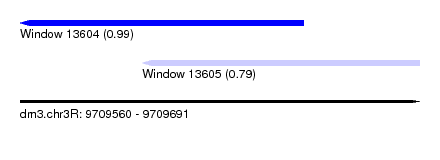
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,709,560 – 9,709,691 |
| Length | 131 |
| Max. P | 0.992300 |

| Location | 9,709,560 – 9,709,653 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 79.69 |
| Shannon entropy | 0.35643 |
| G+C content | 0.45141 |
| Mean single sequence MFE | -29.38 |
| Consensus MFE | -22.13 |
| Energy contribution | -23.05 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.992300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
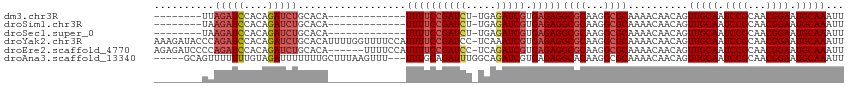
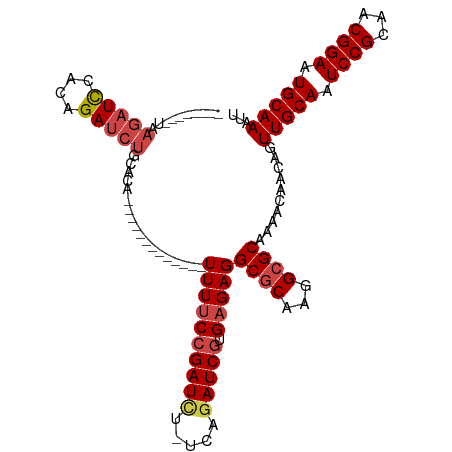
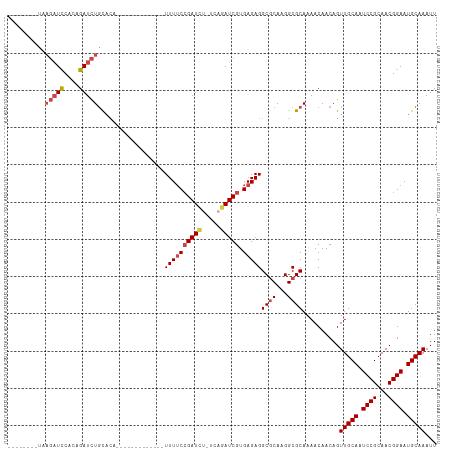
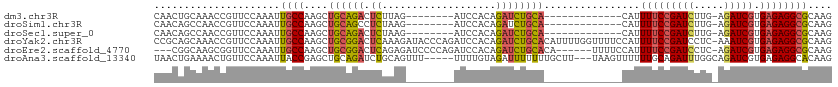
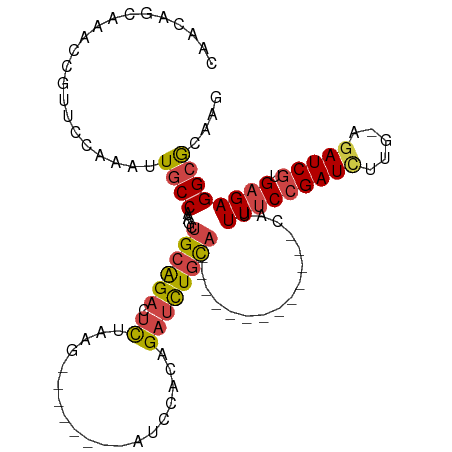
>dm3.chr3R 9709560 93 - 27905053 --------UUAGAUCCACAGAUCUGCACA-------------UUUUCCGAUCU-UGAGAUCGUGAGAGGCGCAAGGCGCAAAACAACAGUUGCAAUCCGCAACGGAAUGCAAAUU --------.((((((....))))))....-------------((((((((((.-...))))).)))))((((...))))..........(((((.((((...)))).)))))... ( -28.90, z-score = -2.89, R) >droSim1.chr3R 15799647 93 + 27517382 --------UAAGAUCCACAGAUCUGCACA-------------UUUUCCGAUCU-UGAGAUCGUGAGAGGCGCAAGGCGCAAAACAACAGUUGCAAUCCGCAACGGAAUGCAAAUU --------..(((((....)))))((...-------------.(((((((((.-...))))).))))(.(....).)))..........(((((.((((...)))).)))))... ( -28.40, z-score = -2.80, R) >droSec1.super_0 8820890 93 + 21120651 --------UAAGAUCCACAGAUCUGCACA-------------UUUUCCGAUCU-UGAGAUCGUGAGAGGCGCAAGGCGCAAAACAACAGUUGCAAUCCGCAACGGAAUGCAAAUU --------..(((((....)))))((...-------------.(((((((((.-...))))).))))(.(....).)))..........(((((.((((...)))).)))))... ( -28.40, z-score = -2.80, R) >droYak2.chr3R 14023815 114 - 28832112 AAAGAUACCCAGAUCCACAGAUCUGCACAUUUUGGUUUUCCAUUUUCCGAUCC-UCAAAUCGUGAGAGGCGCAAGGCGCAAAACAACAGUUGCAAUCCGCAACGGAAUGCAAAUU .(((((...((((((....))))))...))))).((.((((.(((((((((..-....)))).)))))((((...)))).........(((((.....))))))))).))..... ( -30.60, z-score = -2.37, R) >droEre2.scaffold_4770 5824563 108 + 17746568 AGAGAUCCCCAGAUCCACAGAUCUGCACA------UUUUCCAUUUUCCGAUCC-UCAGAUCGUGAGAGGCGCAAGGCGCAAAACAACAGUUGCAAUCCGCAACGGAAUGCAAAUU .((((....((((((....))))))....------.))))..((((((((((.-...))))).)))))((((...))))..........(((((.((((...)))).)))))... ( -31.50, z-score = -3.05, R) >droAna3.scaffold_13340 1064824 107 + 23697760 -----GCAGUUUUUUUGUAGAUUUUUUUGCUUUAAGUUU---UUUGCAGAUUUGGCAGAUCGUGAGAGGCACAAGGCGCAAAACAACAGUUGCAAUCCGCAACGGAAUGCAAAUU -----...(((.(((((((((....)))(((((..((((---((..(.((((.....)))))..))))))..))))))))))).)))..(((((.((((...)))).)))))... ( -28.50, z-score = -1.36, R) >consensus ________UAAGAUCCACAGAUCUGCACA_____________UUUUCCGAUCU_UCAGAUCGUGAGAGGCGCAAGGCGCAAAACAACAGUUGCAAUCCGCAACGGAAUGCAAAUU ..........(((((....)))))..................((((((((((.....))))).)))))((((...))))..........(((((.((((...)))).)))))... (-22.13 = -23.05 + 0.92)
| Location | 9,709,600 – 9,709,691 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 73.43 |
| Shannon entropy | 0.47723 |
| G+C content | 0.48000 |
| Mean single sequence MFE | -26.03 |
| Consensus MFE | -13.23 |
| Energy contribution | -13.51 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.794854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9709600 91 - 27905053 CAACUGCAAACCGUUCCAAAUUGCCAAGCUGCAGACUCUUAG--------AUCCACAGAUCUGCA-------------CAUUUUCCGAUCUUG-AGAUCGUGAGAGGCGCAAG ......................(((..(.((((((.(((...--------......)))))))))-------------)..(((((((((...-.))))).)))))))..... ( -22.30, z-score = -1.15, R) >droSim1.chr3R 15799687 91 + 27517382 CAACAGCCAACCGUUCCAAAUUGCCAAGCUGCAGCCUCUAAG--------AUCCACAGAUCUGCA-------------CAUUUUCCGAUCUUG-AGAUCGUGAGAGGCGCAAG .....((..............(((......)))((((((..(--------(((((.(((((....-------------........)))))))-.))))...))))))))... ( -24.90, z-score = -2.21, R) >droSec1.super_0 8820930 91 + 21120651 CAACAGCCAACCGUUCCAAAUUGCCAAGCUGCAGACUCUAAG--------AUCCACAGAUCUGCA-------------CAUUUUCCGAUCUUG-AGAUCGUGAGAGGCGCAAG .....(((...................(.((((((.(((...--------......)))))))))-------------)..(((((((((...-.))))).)))))))..... ( -22.70, z-score = -1.60, R) >droYak2.chr3R 14023855 112 - 28832112 CCGCAGCAAACCGUUCCAAAUUGCCAAGCUGCGGACUCAAAGAUACCCAGAUCCACAGAUCUGCACAUUUUGGUUUUCCAUUUUCCGAUCCUC-AAAUCGUGAGAGGCGCAAG (((((((....((........))....))))))).(((.........((((((....))))))(((....(((....)))......(((....-..)))))).)))....... ( -27.10, z-score = -1.55, R) >droEre2.scaffold_4770 5824603 103 + 17746568 ---CGGCAAGCGGUUCCAAAUUGCCAAGCUGCGGACUCAGAGAUCCCCAGAUCCACAGAUCUGCACA------UUUUCCAUUUUCCGAUCCUC-AGAUCGUGAGAGGCGCAAG ---.(((((..(....)...)))))....((((..(((.((((....((((((....))))))....------.)))).....(((((((...-.))))).))))).)))).. ( -30.40, z-score = -1.56, R) >droAna3.scaffold_13340 1064864 105 + 23697760 UAACUGAAAACUGUUCCAAAUUACCGAGCUGCAGAUCUGCAGUUU-----UUUUGUAGAUUUUUUUGCUU---UAAGUUUUUUGCAGAUUUGGCAGAUCGUGAGAGGCACAAG ...........(((.((...((((.((.((((((((((((((.((-----(...(((((....)))))..---.)))....)))))))))).)))).))))))..)).))).. ( -28.80, z-score = -2.04, R) >consensus CAACAGCAAACCGUUCCAAAUUGCCAAGCUGCAGACUCUAAG________AUCCACAGAUCUGCA_____________CAUUUUCCGAUCUUG_AGAUCGUGAGAGGCGCAAG .....................((((.....(((((.((...................))))))).................(((((((((.....))))).)))))))).... (-13.23 = -13.51 + 0.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:13:33 2011