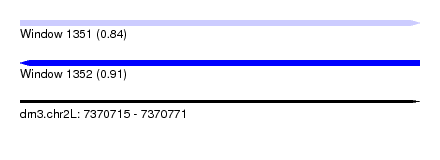
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,370,715 – 7,370,771 |
| Length | 56 |
| Max. P | 0.914769 |

| Location | 7,370,715 – 7,370,771 |
|---|---|
| Length | 56 |
| Sequences | 12 |
| Columns | 59 |
| Reading direction | forward |
| Mean pairwise identity | 84.05 |
| Shannon entropy | 0.36485 |
| G+C content | 0.45733 |
| Mean single sequence MFE | -11.12 |
| Consensus MFE | -5.68 |
| Energy contribution | -6.17 |
| Covariance contribution | 0.49 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.838560 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 7370715 56 + 23011544 CCGAAGCUAUUAACCGUCGCAUUAUUAGACACGACAUGACAC---AAAAGCCACAGCAG .....(((.......((((..((....))..)))).((.(..---....).)).))).. ( -5.00, z-score = -0.57, R) >droSim1.chr2L 7158864 56 + 22036055 CCGAAGCUAUUAACCGUCGCAUUAUUAGACACGACAUGGCAC---AAAAGCCACAGCAG .....(((.......((((..((....))..)))).((((..---....)))).))).. ( -12.10, z-score = -3.11, R) >droSec1.super_3 2885752 56 + 7220098 CCGAAGCUAUUAACCGUCGCAUUAUUAGACACGACAUGGCAC---AAAAGCCACAGCAG .....(((.......((((..((....))..)))).((((..---....)))).))).. ( -12.10, z-score = -3.11, R) >droYak2.chr2L 16791352 56 - 22324452 CCGAAGCUAUUAACUGUCGCAUUAUUAGACACGACAUGGCGC---AAAAGCCACAGCAG .....(((......(((((..((....))..)))))((((..---....)))).))).. ( -13.10, z-score = -2.19, R) >droEre2.scaffold_4929 16285619 55 + 26641161 CCGAAGCUAUUAACCGUCGCAUUAUUAGACACGACAUGGCG----AAAAGCCACAGCAG .....(((.......((((..((....))..)))).((((.----....)))).))).. ( -12.60, z-score = -2.71, R) >droAna3.scaffold_12916 15274142 59 + 16180835 CCGAGGCUAUUAACCGUCGCAUUAUUAGAUGCGACAUGGCAAAAAAAAAGCCACAGCAG .....(((.......((((((((....)))))))).((((.........)))).))).. ( -19.10, z-score = -4.49, R) >dp4.chr4_group3 10976365 56 - 11692001 CCAAAGCUAUUAACCGUCGCAUUAUUAGACACGACAUGGCAU---AAAAGCCACAGCAG .....(((.......((((..((....))..)))).((((..---....)))).))).. ( -12.10, z-score = -3.48, R) >droPer1.super_1 8090974 56 - 10282868 CCAAAGCUAUUAACCGUCGCAUUAUUAGACACGACAUGGCAU---AAAAGCCACAGCAG .....(((.......((((..((....))..)))).((((..---....)))).))).. ( -12.10, z-score = -3.48, R) >droVir3.scaffold_12963 4709950 55 - 20206255 CCGAUGCUAUUAGCCGUCGCAUUAUUAGACAUGCCAUGGCAC----AAAACCACAGCAG ............(((((.((((........)))).)))))..----............. ( -11.40, z-score = -1.38, R) >droMoj3.scaffold_6500 13875576 55 - 32352404 CCGAUGCUAUUAGCCAUCGCAUUAUUAGACACGCCAUGGCAC----AAAACCACAGCAG ............(((((.((............)).)))))..----............. ( -8.90, z-score = -1.15, R) >droGri2.scaffold_15126 4943269 55 + 8399593 CCGAAGCUAUUAGCCGUCGCAUUAUUAGACACGCUAUGGCAG----AAAACCACAGCAG .....(((((.(((.(((.........)))..))))))))..----............. ( -10.70, z-score = -1.25, R) >triCas2.ChLG6 5962876 57 + 13544221 CUAAAUUCAUUAUCAGUUUUAUUAUUAGACAAAUC--GGUAUCUAAUAAACCAUCUCCA .....................((((((((......--....)))))))).......... ( -4.20, z-score = -1.10, R) >consensus CCGAAGCUAUUAACCGUCGCAUUAUUAGACACGACAUGGCAC___AAAAGCCACAGCAG .....(((.......(((.........)))......((((.........)))).))).. ( -5.68 = -6.17 + 0.49)

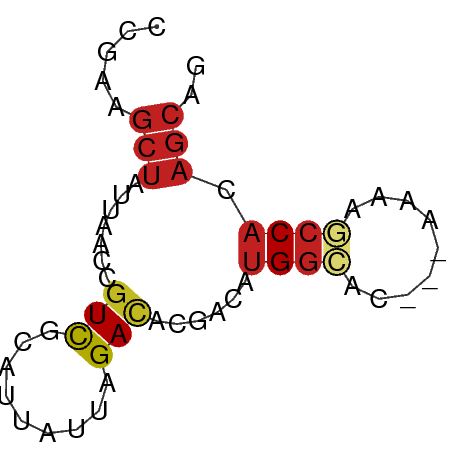
| Location | 7,370,715 – 7,370,771 |
|---|---|
| Length | 56 |
| Sequences | 12 |
| Columns | 59 |
| Reading direction | reverse |
| Mean pairwise identity | 84.05 |
| Shannon entropy | 0.36485 |
| G+C content | 0.45733 |
| Mean single sequence MFE | -16.46 |
| Consensus MFE | -9.45 |
| Energy contribution | -8.82 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.69 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.914769 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 7370715 56 - 23011544 CUGCUGUGGCUUUU---GUGUCAUGUCGUGUCUAAUAAUGCGACGGUUAAUAGCUUCGG ..((((((((....---..))).((((((((......))))))))....)))))..... ( -14.70, z-score = -2.03, R) >droSim1.chr2L 7158864 56 - 22036055 CUGCUGUGGCUUUU---GUGCCAUGUCGUGUCUAAUAAUGCGACGGUUAAUAGCUUCGG ..((((((((....---..))).((((((((......))))))))....)))))..... ( -17.40, z-score = -2.59, R) >droSec1.super_3 2885752 56 - 7220098 CUGCUGUGGCUUUU---GUGCCAUGUCGUGUCUAAUAAUGCGACGGUUAAUAGCUUCGG ..((((((((....---..))).((((((((......))))))))....)))))..... ( -17.40, z-score = -2.59, R) >droYak2.chr2L 16791352 56 + 22324452 CUGCUGUGGCUUUU---GCGCCAUGUCGUGUCUAAUAAUGCGACAGUUAAUAGCUUCGG ..((((((((....---..))).((((((((......))))))))....)))))..... ( -17.50, z-score = -2.24, R) >droEre2.scaffold_4929 16285619 55 - 26641161 CUGCUGUGGCUUUU----CGCCAUGUCGUGUCUAAUAAUGCGACGGUUAAUAGCUUCGG ..((((((((....----.))).((((((((......))))))))....)))))..... ( -16.60, z-score = -2.33, R) >droAna3.scaffold_12916 15274142 59 - 16180835 CUGCUGUGGCUUUUUUUUUGCCAUGUCGCAUCUAAUAAUGCGACGGUUAAUAGCCUCGG ...(((.((((...((...(((..(((((((......)))))))))).)).)))).))) ( -19.70, z-score = -3.48, R) >dp4.chr4_group3 10976365 56 + 11692001 CUGCUGUGGCUUUU---AUGCCAUGUCGUGUCUAAUAAUGCGACGGUUAAUAGCUUUGG ..((((((((....---..))).((((((((......))))))))....)))))..... ( -17.40, z-score = -2.89, R) >droPer1.super_1 8090974 56 + 10282868 CUGCUGUGGCUUUU---AUGCCAUGUCGUGUCUAAUAAUGCGACGGUUAAUAGCUUUGG ..((((((((....---..))).((((((((......))))))))....)))))..... ( -17.40, z-score = -2.89, R) >droVir3.scaffold_12963 4709950 55 + 20206255 CUGCUGUGGUUUU----GUGCCAUGGCAUGUCUAAUAAUGCGACGGCUAAUAGCAUCGG .(((((((((...----..)))))))))............(((..((.....)).))). ( -15.40, z-score = -0.82, R) >droMoj3.scaffold_6500 13875576 55 + 32352404 CUGCUGUGGUUUU----GUGCCAUGGCGUGUCUAAUAAUGCGAUGGCUAAUAGCAUCGG .((((((......----..(((((.((((........)))).)))))..)))))).... ( -18.70, z-score = -2.27, R) >droGri2.scaffold_15126 4943269 55 - 8399593 CUGCUGUGGUUUU----CUGCCAUAGCGUGUCUAAUAAUGCGACGGCUAAUAGCUUCGG ..((((((((...----..)))))))).............(((.(((.....)))))). ( -17.40, z-score = -2.22, R) >triCas2.ChLG6 5962876 57 - 13544221 UGGAGAUGGUUUAUUAGAUACC--GAUUUGUCUAAUAAUAAAACUGAUAAUGAAUUUAG ..........((((((((((..--....)))))))))).....((((........)))) ( -7.90, z-score = -1.08, R) >consensus CUGCUGUGGCUUUU___GUGCCAUGUCGUGUCUAAUAAUGCGACGGUUAAUAGCUUCGG ..((((((((.........)))....(((((......))))).......)))))..... ( -9.45 = -8.82 + -0.64)
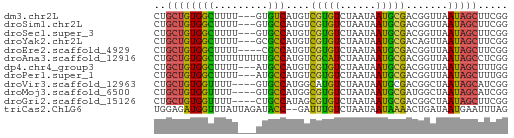
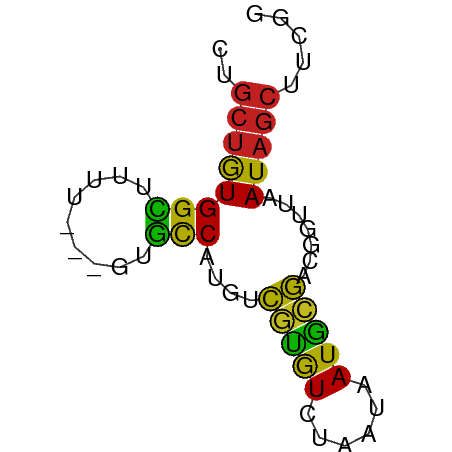
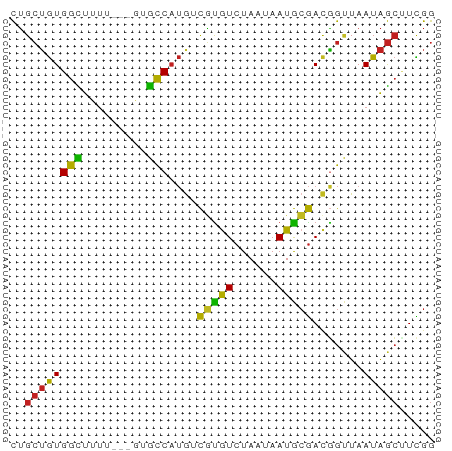
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:23:31 2011