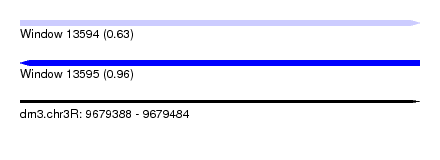
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,679,388 – 9,679,484 |
| Length | 96 |
| Max. P | 0.961795 |

| Location | 9,679,388 – 9,679,484 |
|---|---|
| Length | 96 |
| Sequences | 10 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 78.26 |
| Shannon entropy | 0.45708 |
| G+C content | 0.32255 |
| Mean single sequence MFE | -17.40 |
| Consensus MFE | -12.54 |
| Energy contribution | -12.41 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.12 |
| Mean z-score | -0.84 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.634960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
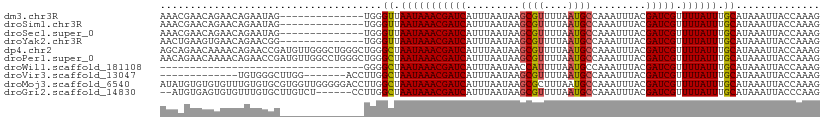
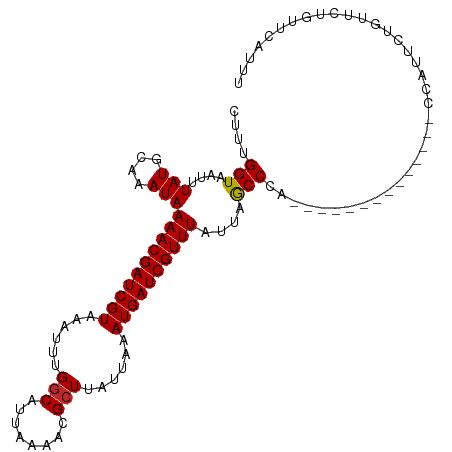
>dm3.chr3R 9679388 96 + 27905053 AAACGAACAGAACAGAAUAG--------------UGGGUUAAUAAACGAUCAUUUAAUAAGCGUUUUAAUGCCAAAUUUACGAUCGUUUUAUUUGCAUAAAUUACCAAAG ...................(--------------..(((....((((((((.........((((....)))).........)))))))).)))..).............. ( -14.47, z-score = -0.91, R) >droSim1.chr3R 15769177 96 - 27517382 AAACGAACAGAACAGAAUAG--------------UGGGUUAAUAAACGAUCAUUUAAUAAGCGUUUUAAUGCCAAAUUUACGAUCGUUUUAUUUGCAUAAAUUACCAAAG ...................(--------------..(((....((((((((.........((((....)))).........)))))))).)))..).............. ( -14.47, z-score = -0.91, R) >droSec1.super_0 8791118 96 - 21120651 AAACGAACAGAACAGAAUAG--------------UGGGUUAAUAAACGAUCAUUUAAUAAGCGUUUUAAUGCCAAAUUUACGAUCGUUUUAUUUGCAUAAAUUACCAAAG ...................(--------------..(((....((((((((.........((((....)))).........)))))))).)))..).............. ( -14.47, z-score = -0.91, R) >droYak2.chr3R 13991933 96 + 28832112 AACUGAAGUGAACAGAACGG--------------UGGGUUAAUAAACGAUCAUUUAAUAAGCGUUUUAAUGCCAAAUUUACGAUCGUUUUAUUUGCAUAAAUUACCAAAG ..(((.......)))...((--------------(((((.(((((((((((.........((((....)))).........))))).)))))).)).....))))).... ( -15.87, z-score = -0.48, R) >dp4.chr2 17065751 110 - 30794189 AGCAGAACAAAACAGAACCGAUGUUGGGCUGGGCUGGGCUAAUAAACGAUCAUUUAAUAAGCGUUUUAAUGCCAAAUUUACGAUCGUUUUAUUUGCAUAAAUUACCAAAG .(((((...............((((((.((.....)).))))))(((((((.........((((....)))).........)))))))...))))).............. ( -20.67, z-score = -0.23, R) >droPer1.super_0 6718633 110 + 11822988 AACAGAACAAAACAGAACCGAUGUUGGCCUGGGCUGGGCUAAUAAACGAUCAUUUAAUAAGCGUUUUAAUGCCAAAUUUACGAUCGUUUUAUUUGCAUAAAUUACCAAAG ............((((.....(((((((((.....)))))))))(((((((.........((((....)))).........)))))))...))))............... ( -20.87, z-score = -0.76, R) >droWil1.scaffold_181108 1810709 76 + 4707319 ----------------------------------GGGGCUAAUAAACGAUCAUUUAAUAACCAUUUUAAUGCCAAAUUUACGAUCGUUUUAUUUGCAUAAAUUACCAAAG ----------------------------------((.((.(((((((((((..........(((....)))..........))))).)))))).))........)).... ( -10.55, z-score = -1.13, R) >droVir3.scaffold_13047 422180 90 - 19223366 -------------UGUGGGCUUGG-------ACCUUGGCUAAUAAACGAUCAUUUAAUAAGCGUUUUAAUGCCAAAUUUACGAUCGUUUUAUUUGCAUAAAUUACCAAAG -------------.......((((-------......((.(((((((((((.........((((....)))).........))))).)))))).))........)))).. ( -15.81, z-score = -0.48, R) >droMoj3.scaffold_6540 19607633 110 + 34148556 AUAUGUGUGUGUUUGUGUGCGUGGUUGGGGGACCUUGGCUAAUAAACGAUCAUUUAAUAAGCGCUUUAAUGCCAAAUUUACGAUCGUUUUAUUUGCAUAAAUUACCAAAG ...((.(((.((((((((...((((..((....))..))))..((((((((....(((..(.((......)))..)))...)))))))).....)))))))))))))... ( -25.40, z-score = -0.57, R) >droGri2.scaffold_14830 2335433 102 - 6267026 --AUGUGAGUGUGUUUGUGCUUGUCU------CCUUGGCUAAUAAACGAUCAUUUAAUAAGCGUUUUAAUGCCAAAUUUACGAUCGUUUUAUUUGCAUAAAUUACCCAAG --...((.(((.((((((((..(((.------....)))....((((((((.........((((....)))).........)))))))).....))))))))))).)).. ( -21.37, z-score = -2.05, R) >consensus AAACGAACAGAACAGAAUGG______________UGGGCUAAUAAACGAUCAUUUAAUAAGCGUUUUAAUGCCAAAUUUACGAUCGUUUUAUUUGCAUAAAUUACCAAAG .....................................((.(((((((((((.........((((....)))).........))))).)))))).)).............. (-12.54 = -12.41 + -0.13)

| Location | 9,679,388 – 9,679,484 |
|---|---|
| Length | 96 |
| Sequences | 10 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 78.26 |
| Shannon entropy | 0.45708 |
| G+C content | 0.32255 |
| Mean single sequence MFE | -18.00 |
| Consensus MFE | -14.57 |
| Energy contribution | -14.43 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.961795 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
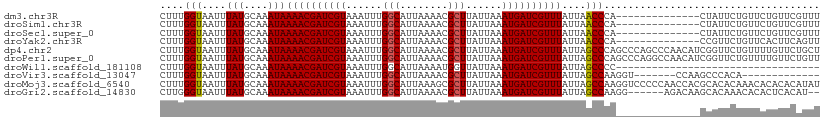
>dm3.chr3R 9679388 96 - 27905053 CUUUGGUAAUUUAUGCAAAUAAAACGAUCGUAAAUUUGGCAUUAAAACGCUUAUUAAAUGAUCGUUUAUUAACCCA--------------CUAUUCUGUUCUGUUCGUUU ....(((....(((....)))((((((((((......(((........)))......))))))))))....)))..--------------.................... ( -14.50, z-score = -0.87, R) >droSim1.chr3R 15769177 96 + 27517382 CUUUGGUAAUUUAUGCAAAUAAAACGAUCGUAAAUUUGGCAUUAAAACGCUUAUUAAAUGAUCGUUUAUUAACCCA--------------CUAUUCUGUUCUGUUCGUUU ....(((....(((....)))((((((((((......(((........)))......))))))))))....)))..--------------.................... ( -14.50, z-score = -0.87, R) >droSec1.super_0 8791118 96 + 21120651 CUUUGGUAAUUUAUGCAAAUAAAACGAUCGUAAAUUUGGCAUUAAAACGCUUAUUAAAUGAUCGUUUAUUAACCCA--------------CUAUUCUGUUCUGUUCGUUU ....(((....(((....)))((((((((((......(((........)))......))))))))))....)))..--------------.................... ( -14.50, z-score = -0.87, R) >droYak2.chr3R 13991933 96 - 28832112 CUUUGGUAAUUUAUGCAAAUAAAACGAUCGUAAAUUUGGCAUUAAAACGCUUAUUAAAUGAUCGUUUAUUAACCCA--------------CCGUUCUGUUCACUUCAGUU ....(((....(((....)))((((((((((......(((........)))......))))))))))........)--------------))...(((.......))).. ( -15.90, z-score = -1.25, R) >dp4.chr2 17065751 110 + 30794189 CUUUGGUAAUUUAUGCAAAUAAAACGAUCGUAAAUUUGGCAUUAAAACGCUUAUUAAAUGAUCGUUUAUUAGCCCAGCCCAGCCCAACAUCGGUUCUGUUUUGUUCUGCU ..............((.....((((((((((......(((........)))......))))))))))...((..(((..(((.((......))..)))..)))..)))). ( -21.30, z-score = -0.86, R) >droPer1.super_0 6718633 110 - 11822988 CUUUGGUAAUUUAUGCAAAUAAAACGAUCGUAAAUUUGGCAUUAAAACGCUUAUUAAAUGAUCGUUUAUUAGCCCAGCCCAGGCCAACAUCGGUUCUGUUUUGUUCUGUU .....................((((((((((......(((........)))......))))))))))..(((..(((..((((((......)).))))..)))..))).. ( -22.90, z-score = -1.12, R) >droWil1.scaffold_181108 1810709 76 - 4707319 CUUUGGUAAUUUAUGCAAAUAAAACGAUCGUAAAUUUGGCAUUAAAAUGGUUAUUAAAUGAUCGUUUAUUAGCCCC---------------------------------- ..............((.((((((.(((((((......(.(((....))).)......))))))))))))).))...---------------------------------- ( -13.20, z-score = -0.95, R) >droVir3.scaffold_13047 422180 90 + 19223366 CUUUGGUAAUUUAUGCAAAUAAAACGAUCGUAAAUUUGGCAUUAAAACGCUUAUUAAAUGAUCGUUUAUUAGCCAAGGU-------CCAAGCCCACA------------- (((((((....(((....)))((((((((((......(((........)))......))))))))))....))))))).-------...........------------- ( -20.30, z-score = -2.40, R) >droMoj3.scaffold_6540 19607633 110 - 34148556 CUUUGGUAAUUUAUGCAAAUAAAACGAUCGUAAAUUUGGCAUUAAAGCGCUUAUUAAAUGAUCGUUUAUUAGCCAAGGUCCCCCAACCACGCACACAAACACACACAUAU (((((((....(((....)))((((((((((......(((........)))......))))))))))....)))))))................................ ( -20.30, z-score = -1.62, R) >droGri2.scaffold_14830 2335433 102 + 6267026 CUUGGGUAAUUUAUGCAAAUAAAACGAUCGUAAAUUUGGCAUUAAAACGCUUAUUAAAUGAUCGUUUAUUAGCCAAGG------AGACAAGCACAAACACACUCACAU-- ..(((((..(((.(((.....((((((((((......(((........)))......)))))))))).........(.------...)..))).)))...)))))...-- ( -22.60, z-score = -3.35, R) >consensus CUUUGGUAAUUUAUGCAAAUAAAACGAUCGUAAAUUUGGCAUUAAAACGCUUAUUAAAUGAUCGUUUAUUAGCCCA______________CCAUUCUGUUCUGUUCAUUU ....(((....(((....)))((((((((((......(((........)))......))))))))))....))).................................... (-14.57 = -14.43 + -0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:13:25 2011