
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,670,846 – 9,670,960 |
| Length | 114 |
| Max. P | 0.782971 |

| Location | 9,670,846 – 9,670,960 |
|---|---|
| Length | 114 |
| Sequences | 11 |
| Columns | 128 |
| Reading direction | forward |
| Mean pairwise identity | 76.96 |
| Shannon entropy | 0.48474 |
| G+C content | 0.49451 |
| Mean single sequence MFE | -36.45 |
| Consensus MFE | -14.92 |
| Energy contribution | -14.73 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.755589 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
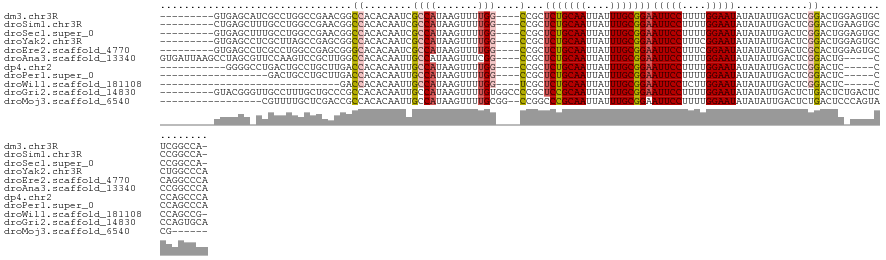
>dm3.chr3R 9670846 114 + 27905053 -UGGCCGAGCACUCCAGUCCGAGUCAAUAUAUAUUCCAAAAGGAAUUCCGCAAAUAAUUGCAGAGCGG----CCAAAACUUAUGGCGAUUGUGUGGCCGUUCGGCCAGGCGAUGCUCAC--------- -((((((..(......)..)).))))......(((((....)))))...((((....)))).((((.(----(((.......)))).(((((.(((((....))))).)))))))))..--------- ( -40.50, z-score = -2.70, R) >droSim1.chr3R 15760727 114 - 27517382 -UGGCCGGGCACUUCAGUCCGAGUCAAUAUAUAUUCCAAAAGGAAUUCCGCAAAUAAUUGCAGAGCGG----CCAAAACUUAUGGCGAUUGUGUGGCCGUUCGGCCAGGCAAAGCUCAG--------- -(((((((((......))))).))))......(((((....)))))...((((....)))).((((.(----(((.......))))..((((.(((((....))))).)))).))))..--------- ( -45.20, z-score = -4.21, R) >droSec1.super_0 8782696 114 - 21120651 -UGGCCGGGCACUCCAGUCCGAGUCAAUAUAUAUUCCAAAAGGAAUUCCGCAAAUAAUUGCAGAGCGG----CCAAAACUUAUGGCGAUUGUGUGGCCGUUCGGCCAGGCAAAGCUCAC--------- -(((((((((......))))).))))......(((((....)))))...((((....)))).((((.(----(((.......))))..((((.(((((....))))).)))).))))..--------- ( -45.20, z-score = -4.04, R) >droYak2.chr3R 13983094 115 + 28832112 UGGGCCAGGCACUCCAGUCCGAGUCAAUAUAUAUUCCGAAAGGAAUUCCGCAAAUAAUUGCAGAGCGG----CCAAAACUUAUGGCGAUUGUGUGGCCGCUCGGCUAAGCGAGGCUCAC--------- ((((((.((.((....)))).((((.......(((((....)))))...((((....)))).((((((----(((..((...........)).)))))))))))))......)))))).--------- ( -44.40, z-score = -3.12, R) >droEre2.scaffold_4770 5784629 115 - 17746568 UGGGCCUGGCACUCCAGUGCGAGUCAAUAUAUAUUCCGAAAGGAAUUCCGCAAAUAAUUGCAGAGCGG----CCAAAACUUAUGGCGAUUGUGUGCCCGCUCGGCCAGGCGAGGCUCAC--------- ((((((((((((..((((((............(((((....)))))((.((((....)))).)))).(----(((.......)))).)))).)))))((((......))))))))))).--------- ( -42.80, z-score = -1.94, R) >droAna3.scaffold_13340 1028309 119 - 23697760 UGGGCCGGG-----CAGUCCGAGUCAAUAUAUAUUCCAAAAGGAAUUCCGCAAAUAAUUGCAGAGCGG----CCGAAACUUAUGGCAAUUGUGUGGCCAAGCGGACUUGGAACGCUAGGCUUAAUCAC ((((((.((-----(..((((((((.......(((((....)))))..(((........((...))((----(((..((...........)).)))))..)))))))))))..))).))))))..... ( -41.40, z-score = -2.60, R) >dp4.chr2 17057533 108 - 30794189 UGGGCUGGG-----GAGUCCGAGUCAAUAUAUAUUCCAAAAGGAAUUCCGCAAAUAAUUGCAGAGCGG----CCAAAACUUAUGGCAAUUGUGUGGUCAAGCAGGCAGUCAGGCCCC----------- .(((((...-----((.(((............(((((....)))))...((.....((..((.((..(----(((.......))))..)).))..))...)).)).).)).))))).----------- ( -29.30, z-score = 0.06, R) >droPer1.super_0 6710397 101 + 11822988 UGGGCUGGG-----GAGUCCGAGUCAAUAUAUAUUCCAAAAGGAAUUCCGCAAAUAAUUGCAGAGCGG----CCAAAACUUAUGGCAAUUGUGUGGUCAAGCAGGCAGUC------------------ .(((((...-----.)))))..(((.......(((((....)))))...((.....((..((.((..(----(((.......))))..)).))..))...)).)))....------------------ ( -25.00, z-score = -0.20, R) >droWil1.scaffold_181108 1802954 88 + 4707319 -CGGCUGGG-----GAGUCCGAGUCAAUAUAUAUUCCAAGAGGAAUUCCGCAAAUAAUUGCAGAGCGA----CCAAAACUUAUGGCAAUUGUGUGGUC------------------------------ -.((((.((-----....)).)))).......(((((....))))).(((((..(((((((.(((...----......)))...))))))))))))..------------------------------ ( -22.80, z-score = -1.12, R) >droGri2.scaffold_14830 2326986 119 - 6267026 UGCACUGGGAGUCAGAGUCAGAGUCAAUAUAUAUUCCAAAAGGAAUUCCGCAAAUAAUUGCGGAGCGGGGCCACAAAACUUAUGGCAAUUGUGUGGCGGGCAGCAAAGGCAACCCGUAC--------- ........((.((.......)).)).......(((((....)))))(((((((....)))))))(((((((((((...(....).......)))))).....((....))..)))))..--------- ( -34.10, z-score = -1.10, R) >droMoj3.scaffold_6540 19597387 103 + 34148556 ------CGUACUGGGAGUCAGAGUCAAUAUAUAUUCCAAAAGGAAUUCCGCAAAUAAUUGCGGGCCGG--CCGCAAAACUUAUGGCAAUUGUGUGGCGGUCGAGCAAAACG----------------- ------....(((.....)))...........(((((....))))).((((((....))))))(((((--((((...((...........))...))))))).))......----------------- ( -30.20, z-score = -1.59, R) >consensus UGGGCCGGGCACU_CAGUCCGAGUCAAUAUAUAUUCCAAAAGGAAUUCCGCAAAUAAUUGCAGAGCGG____CCAAAACUUAUGGCAAUUGUGUGGCCGUUCGGCCAGGCAA_GCUCAC_________ ..((((..........................(((((....))))).......((((((((.(((.............)))...))))))))..)))).............................. (-14.92 = -14.73 + -0.19)


| Location | 9,670,846 – 9,670,960 |
|---|---|
| Length | 114 |
| Sequences | 11 |
| Columns | 128 |
| Reading direction | reverse |
| Mean pairwise identity | 76.96 |
| Shannon entropy | 0.48474 |
| G+C content | 0.49451 |
| Mean single sequence MFE | -33.60 |
| Consensus MFE | -14.58 |
| Energy contribution | -14.80 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.782971 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9670846 114 - 27905053 ---------GUGAGCAUCGCCUGGCCGAACGGCCACACAAUCGCCAUAAGUUUUGG----CCGCUCUGCAAUUAUUUGCGGAAUUCCUUUUGGAAUAUAUAUUGACUCGGACUGGAGUGCUCGGCCA- ---------.(((((((..((...((((.((((((.((...........))..)))----))).(((((((....)))))))(((((....)))))..........))))...)).)))))))....- ( -39.10, z-score = -2.01, R) >droSim1.chr3R 15760727 114 + 27517382 ---------CUGAGCUUUGCCUGGCCGAACGGCCACACAAUCGCCAUAAGUUUUGG----CCGCUCUGCAAUUAUUUGCGGAAUUCCUUUUGGAAUAUAUAUUGACUCGGACUGAAGUGCCCGGCCA- ---------.((.((.(((..(((((....)))))..)))..)))).......(((----(((.(((((((....)))))))(((((....)))))............((((....)).))))))))- ( -36.60, z-score = -2.01, R) >droSec1.super_0 8782696 114 + 21120651 ---------GUGAGCUUUGCCUGGCCGAACGGCCACACAAUCGCCAUAAGUUUUGG----CCGCUCUGCAAUUAUUUGCGGAAUUCCUUUUGGAAUAUAUAUUGACUCGGACUGGAGUGCCCGGCCA- ---------(((.((.(((..(((((....)))))..)))..)))))......(((----(((.(((((((....)))))))(((((....)))))........((((......))))...))))))- ( -40.20, z-score = -2.44, R) >droYak2.chr3R 13983094 115 - 28832112 ---------GUGAGCCUCGCUUAGCCGAGCGGCCACACAAUCGCCAUAAGUUUUGG----CCGCUCUGCAAUUAUUUGCGGAAUUCCUUUCGGAAUAUAUAUUGACUCGGACUGGAGUGCCUGGCCCA ---------.((.(((.((((((((((((((((((.((...........))..)))----))))(((((((....)))))))(((((....)))))..........)))).)).)))))...))).)) ( -40.80, z-score = -2.33, R) >droEre2.scaffold_4770 5784629 115 + 17746568 ---------GUGAGCCUCGCCUGGCCGAGCGGGCACACAAUCGCCAUAAGUUUUGG----CCGCUCUGCAAUUAUUUGCGGAAUUCCUUUCGGAAUAUAUAUUGACUCGCACUGGAGUGCCAGGCCCA ---------.((.((((.((((((((((((.(((........)))....).)))))----))(((((((((....)))))))(((((....)))))............)).....)).)).)))).)) ( -39.90, z-score = -1.32, R) >droAna3.scaffold_13340 1028309 119 + 23697760 GUGAUUAAGCCUAGCGUUCCAAGUCCGCUUGGCCACACAAUUGCCAUAAGUUUCGG----CCGCUCUGCAAUUAUUUGCGGAAUUCCUUUUGGAAUAUAUAUUGACUCGGACUG-----CCCGGCCCA ........(((..(((.(((.((((.((..((((....((((......))))..))----))))(((((((....)))))))(((((....))))).......)))).))).))-----)..)))... ( -36.80, z-score = -2.16, R) >dp4.chr2 17057533 108 + 30794189 -----------GGGGCCUGACUGCCUGCUUGACCACACAAUUGCCAUAAGUUUUGG----CCGCUCUGCAAUUAUUUGCGGAAUUCCUUUUGGAAUAUAUAUUGACUCGGACUC-----CCCAGCCCA -----------((((((.((((....(((((......)))..))....))))..))----))(((((((((....)))))))(((((....)))))............((....-----.)).)))). ( -29.10, z-score = -1.21, R) >droPer1.super_0 6710397 101 - 11822988 ------------------GACUGCCUGCUUGACCACACAAUUGCCAUAAGUUUUGG----CCGCUCUGCAAUUAUUUGCGGAAUUCCUUUUGGAAUAUAUAUUGACUCGGACUC-----CCCAGCCCA ------------------........(((........((((.((((.......)))----)...(((((((....)))))))(((((....)))))....))))....((....-----.)))))... ( -20.90, z-score = -0.72, R) >droWil1.scaffold_181108 1802954 88 - 4707319 ------------------------------GACCACACAAUUGCCAUAAGUUUUGG----UCGCUCUGCAAUUAUUUGCGGAAUUCCUCUUGGAAUAUAUAUUGACUCGGACUC-----CCCAGCCG- ------------------------------............((....((((..((----((..(((((((....)))))))(((((....))))).......))))..)))).-----....))..- ( -22.10, z-score = -2.27, R) >droGri2.scaffold_14830 2326986 119 + 6267026 ---------GUACGGGUUGCCUUUGCUGCCCGCCACACAAUUGCCAUAAGUUUUGUGGCCCCGCUCCGCAAUUAUUUGCGGAAUUCCUUUUGGAAUAUAUAUUGACUCUGACUCUGACUCCCAGUGCA ---------((((((((.((....)).))))((((((.((((......)))).)))))).....(((((((....)))))))(((((....)))))...........................)))). ( -38.90, z-score = -3.91, R) >droMoj3.scaffold_6540 19597387 103 - 34148556 -----------------CGUUUUGCUCGACCGCCACACAAUUGCCAUAAGUUUUGCGG--CCGGCCCGCAAUUAUUUGCGGAAUUCCUUUUGGAAUAUAUAUUGACUCUGACUCCCAGUACG------ -----------------.(((....(((((((((.((.((((......)))).)).))--.))).((((((....)))))).(((((....))))).....))))....)))..........------ ( -25.20, z-score = -1.53, R) >consensus _________GUGAGC_UUGCCUGGCCGAACGGCCACACAAUUGCCAUAAGUUUUGG____CCGCUCUGCAAUUAUUUGCGGAAUUCCUUUUGGAAUAUAUAUUGACUCGGACUG_AGUGCCCGGCCCA ................................................((((..((........(((((((....)))))))(((((....))))).........))..))))............... (-14.58 = -14.80 + 0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:13:23 2011