| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,668,679 – 9,668,821 |
| Length | 142 |
| Max. P | 0.724973 |

| Location | 9,668,679 – 9,668,821 |
|---|---|
| Length | 142 |
| Sequences | 5 |
| Columns | 173 |
| Reading direction | forward |
| Mean pairwise identity | 74.47 |
| Shannon entropy | 0.41266 |
| G+C content | 0.47305 |
| Mean single sequence MFE | -38.20 |
| Consensus MFE | -19.82 |
| Energy contribution | -20.14 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.724973 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
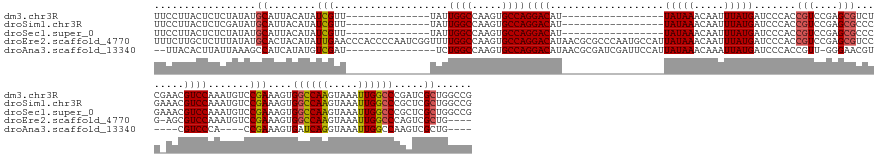
>dm3.chr3R 9668679 142 + 27905053 UUCCUUACUCUCUAUAUGCAUUACAUAUCGUU--------------UAUUGGCCAAGUGCCAGGACAU-----------------UAUAAACAAUUUAUGAUCCCACCGUCCGAGCGUCUCGAACGUCCAAAUGUCCGAAAGUGGCCAAGUAAAUUGGCCCGAUCGCUGGCCG .............(((((.....)))))....--------------....(((((.((....((((((-----------------(((((.....))))))......(((.((((...)))).)))......)))))((..(.((((((.....)))))))..))))))))). ( -36.80, z-score = -2.02, R) >droSim1.chr3R 15758563 142 - 27517382 UUCCUUACUCUCGAUAUGCAUUACAUAUCGUU--------------UAUUGGCCAAGUGCCAGGACAU-----------------UAUAAACAAUUUAUGAUCCCACCGUCCGAGCGCCCGAAACGUCCAAAUGUCCGAAAGUGGCCAAGUAAAUUGGCCCGCUCGCUGGCCG ...........(((((((.....)))))))..--------------....(((((.((....((((((-----------------(((((.....)))))))......))))(((((.......((..(....)..)).....((((((.....)))))))))))))))))). ( -42.30, z-score = -3.25, R) >droSec1.super_0 8780537 142 - 21120651 UUCCUUACUCUCUAUAUGCAUUACAUAUCGUU--------------UAUUGGCCAAGUGCCAGGACAU-----------------UAUAAACAAUUUAUGAUCCCACCGUCCGAGCGCCCGAAACGUCCAAAUGUCCGAAAGUGGCCAAGUAAAUUGGCCCGCUCGCUGGCCG .............(((((.....)))))....--------------....(((((.((....((((((-----------------(((((.....)))))))......))))(((((.......((..(....)..)).....((((((.....)))))))))))))))))). ( -36.60, z-score = -1.92, R) >droEre2.scaffold_4770 5782078 168 - 17746568 UUUCUUGCUCUUUAUAUGCACUACAUAUUGAACCCACCCCAAUCGGUUUUGGCCAAGUGCCAGGACAUAACGCGCCCAAUGCCAUUAUAAACAAUUUAUGAUCCCACCGUCCGAGCGUCCG-AGCGUCCAAAUGUCCGAAAGUGGCCAAGUAAAUUGGCCCAGUCGCUG---- ......((((.......(((((.......(((((..........)))))......)))))..((((.....(((.....))).(((((((.....)))))))......)))))))).....-((((.(........(....).((((((.....))))))..).)))).---- ( -39.52, z-score = -0.84, R) >droAna3.scaffold_13340 1026108 143 - 23697760 --UUACACUUAUUAAAGCCAUCAUAUGUCGAU---------------UCUGGCCAAGUGCCAGGACAUAACGCGAUCGAUUCCAUUAUAAACAAAUUAUGAUCCCACCGUU-GGGAACGU----CGUCCCA----CCGAAAGUGAUCAGGUAAAUUGGCCAAGUCGCUG---- --........................(.((((---------------(.(((((((.((((.((((((((.(.((.....))).)))).........(((.(((((....)-)))).)))----.))))((----.(....)))....))))..)))))))))))))..---- ( -35.80, z-score = -1.34, R) >consensus UUCCUUACUCUCUAUAUGCAUUACAUAUCGUU______________UAUUGGCCAAGUGCCAGGACAU_________________UAUAAACAAUUUAUGAUCCCACCGUCCGAGCGCCCG_AACGUCCAAAUGUCCGAAAGUGGCCAAGUAAAUUGGCCCGCUCGCUGGCCG .................((........(((...................((((.....))))((((...................(((((.....))))).......(((....)))........)))).......)))....((((((.....)))))).....))...... (-19.82 = -20.14 + 0.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:13:21 2011