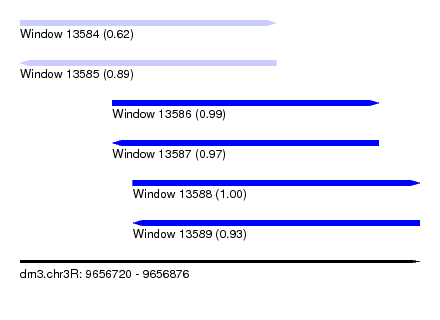
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,656,720 – 9,656,876 |
| Length | 156 |
| Max. P | 0.995911 |

| Location | 9,656,720 – 9,656,820 |
|---|---|
| Length | 100 |
| Sequences | 12 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 81.22 |
| Shannon entropy | 0.36790 |
| G+C content | 0.40316 |
| Mean single sequence MFE | -28.34 |
| Consensus MFE | -16.19 |
| Energy contribution | -16.94 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.620844 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9656720 100 + 27905053 --UGCUAGUUGAGCUUUGGAAAAACCAUGAAAUGUGCG--AGUA-CGAG-----------GAAAAUCUUCCGCGUUUGUGUAAUGCAGGAAGAUUUU--CCAUGAUGUUU-G-UUUUGUA- --.(((.....)))..(((.....)))........(((--((((-((.(-----------((((((((((((((((.....))))).))))))))))--)).)).)))))-)-)......- ( -28.40, z-score = -2.82, R) >droSim1.chr3R 15746893 100 - 27517382 --UGCUAGUUGAGCUCUGGAAAAACCAUGAAAUGUGCG--AGUA-AGAG-----------GAAAAUCUUCCGCGUUUGUGUAAUGCAGGAAGAUUUU--CCAUGAUGUUU-G-UUUUGUA- --((((.((.....(((((.....))).)).....)).--))))-...(-----------((((((((((((((((.....))))).))))))))))--)).........-.-.......- ( -29.10, z-score = -3.25, R) >droSec1.super_0 8769352 100 - 21120651 --UGCUAGUUGAGCUCUGGAAAAACCAUGAAAUGUGCG--AGUA-AGAG-----------GAAAAUCUUCCGCGUUUGUGUAAUGCAGGAAGAUUUU--CCACGUUGUUU-G-UUUUCUA- --.(((.....)))..(((((((..((...(((((.(.--....-.).(-----------((((((((((((((((.....))))).))))))))))--)))))))...)-)-)))))))- ( -30.00, z-score = -3.01, R) >droYak2.chr3R 13967057 100 + 28832112 --GGCAGGUUGAGCUCUGGAAAAUCCAUGAAAUGUGCG--AGUA-CGAG-----------GAAAAUCUUCCGCGUUUGUGUAAUGCAGGAAGAUUUU--CCAUGAUGUUU-G-UUUUGUA- --.(((((..(((((((((.....))).))..((((..--..))-)).(-----------((((((((((((((((.....))))).))))))))))--)).....))))-.-.))))).- ( -32.50, z-score = -3.28, R) >droEre2.scaffold_4770 5771147 99 - 17746568 --UGCAGGUUGAGCUGUGGAAAACCCAUGAAAUGUGCG--AGUA-CGAG-----------GAAAAU-UUCCGCGUUUGUGUAAUGCAGGAAGAUUUU--CCAUGAUGUUU-G-UUUUGUA- --((((((..((((.((((.....))))....((((..--..))-)).(-----------((((((-(((((((((.....))))).)).)))))))--)).....))))-.-.))))))- ( -25.20, z-score = -1.38, R) >droAna3.scaffold_13340 17983247 101 + 23697760 --UGCAGGUUGAGCUCUGAAAAAUCCAUGAAACAAGCA--AGUA-CGGU-----------GAAAAUCUUCCGCGUUUGUGUAAUGCAGGAAGAUUUU--GCAUGAUUUUC-GCUUUUGUG- --.((((...((((...(((((...((((..((.....--.)).-....-----------.(((((((((((((((.....))))).))))))))))--.)))).)))))-)))))))).- ( -23.90, z-score = -0.26, R) >dp4.chr2 15651228 101 + 30794189 --UGCAGUUGGAGCGCUGGAAAAUCCAUGAAAUGUGCG--AGUA-CAGC-----------GAAAACCUUCCGCGUUUGUGUAAUGCAGGAAGAUUUU--GCAUGAUGUUU-GCCUUUGUC- --(((((((....(((((.....(((((.....))).)--)...-))))-----------)..)))((((((((((.....))))).)))))....)--)))........-.........- ( -25.90, z-score = -0.26, R) >droPer1.super_0 7061094 101 - 11822988 --UGCAGUUGGAGCGCUGGAAAAUCCAUGAAAUGUGCG--AGUA-CAGC-----------GAAAACCUUCCGCGUUUGUGUAAUGCAGGAAGAUUUU--GCAUGAUGUUU-GCCUUUGUC- --(((((((....(((((.....(((((.....))).)--)...-))))-----------)..)))((((((((((.....))))).)))))....)--)))........-.........- ( -25.90, z-score = -0.26, R) >droWil1.scaffold_181130 11123110 105 + 16660200 UGUAGUAGUUGGGCACUGGAAAAAUCAUGAAAUGUGCGCAAGUA-UAUG-----------GCAAAUCUUCCGCGUUUGUGUAAUGCAGGAAGAUUUUU-GCAUGAUAUUU-G-CUUUUUG- ............((((.................))))(((((((-(...-----------((((((((((((((((.....))))).)))))))..))-))...))))))-)-)......- ( -31.83, z-score = -2.62, R) >droVir3.scaffold_12822 1648891 110 + 4096053 --UGCAG--UCAGCACUGGAAAAUCCAUGAAAUGUGCG--AGUA-CAAGUAUUAAAGCAAGAAAAUCUUCCGCGUUUGUGUAAUGCAGGAAGAUUUU--GCAUGAUGUUU-GCUUUUAUU- --.((((--(((((..(((.....))).......(((.--((((-....))))...)))..(((((((((((((((.....))))).))))))))))--)).))))...)-)).......- ( -29.30, z-score = -1.70, R) >droMoj3.scaffold_6540 22685027 111 - 34148556 --UGCAG--UCAACUUUGGGAAAUCCAUGAAAUGUGCG--AGUA-CAAGUAUUAAAGCAAGCAAAUCUUCCGCGUUUGUGUAAUGCAGGAAGAUUUU--GCAUGAUUUUUCGCUUUUAUU- --.((((--....))..(..(((((.........(((.--((((-....))))...))).((((((((((((((((.....))))).)))))).)))--))..)))))..))).......- ( -29.50, z-score = -1.70, R) >droGri2.scaffold_14624 2694103 112 + 4233967 --UGCUG--UUAGUACUGGCAAAUCCAUGAAAUGUGCG--AGUAGCAAGCAUUAAAGCAAGAAAAUCUUCCGCGUUUGUGUAAUUCAGGAAGAUUUUUUGCACGAUGUUG---UUUUGCAU --(((((--(....)).))))............(((((--((..((((.((((...((((((((.((((((..(((.....)))...))))))))))))))..)))))))---)))))))) ( -28.50, z-score = -0.81, R) >consensus __UGCAGGUUGAGCUCUGGAAAAUCCAUGAAAUGUGCG__AGUA_CAAG___________GAAAAUCUUCCGCGUUUGUGUAAUGCAGGAAGAUUUU__GCAUGAUGUUU_G_UUUUGUA_ .........................((((.....(((....))).................(((((((((((((((.....))))).))))))))))...))))................. (-16.19 = -16.94 + 0.75)

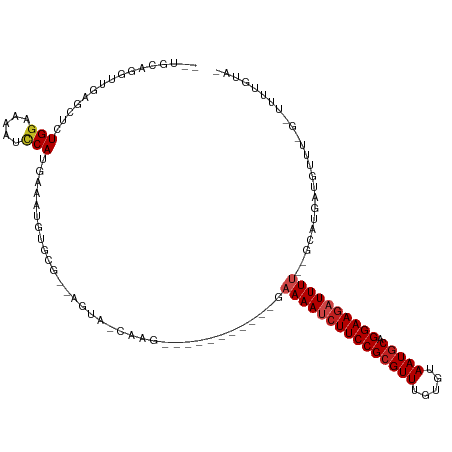
| Location | 9,656,720 – 9,656,820 |
|---|---|
| Length | 100 |
| Sequences | 12 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 81.22 |
| Shannon entropy | 0.36790 |
| G+C content | 0.40316 |
| Mean single sequence MFE | -21.95 |
| Consensus MFE | -11.55 |
| Energy contribution | -11.76 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.885549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9656720 100 - 27905053 -UACAAAA-C-AAACAUCAUGG--AAAAUCUUCCUGCAUUACACAAACGCGGAAGAUUUUC-----------CUCG-UACU--CGCACAUUUCAUGGUUUUUCCAAAGCUCAACUAGCA-- -.......-.-.........((--((((((((((.((...........)))))))))))))-----------)...-....--...........(((.....)))..(((.....))).-- ( -22.40, z-score = -4.21, R) >droSim1.chr3R 15746893 100 + 27517382 -UACAAAA-C-AAACAUCAUGG--AAAAUCUUCCUGCAUUACACAAACGCGGAAGAUUUUC-----------CUCU-UACU--CGCACAUUUCAUGGUUUUUCCAGAGCUCAACUAGCA-- -.......-.-.........((--((((((((((.((...........)))))))))))))-----------)...-....--........((.(((.....)))))(((.....))).-- ( -23.00, z-score = -4.36, R) >droSec1.super_0 8769352 100 + 21120651 -UAGAAAA-C-AAACAACGUGG--AAAAUCUUCCUGCAUUACACAAACGCGGAAGAUUUUC-----------CUCU-UACU--CGCACAUUUCAUGGUUUUUCCAGAGCUCAACUAGCA-- -.......-.-.......(.((--((((((((((.((...........)))))))))))))-----------).).-....--........((.(((.....)))))(((.....))).-- ( -23.60, z-score = -3.30, R) >droYak2.chr3R 13967057 100 - 28832112 -UACAAAA-C-AAACAUCAUGG--AAAAUCUUCCUGCAUUACACAAACGCGGAAGAUUUUC-----------CUCG-UACU--CGCACAUUUCAUGGAUUUUCCAGAGCUCAACCUGCC-- -.......-.-.........((--((((((((((.((...........)))))))))))))-----------)...-....--.(((...(((.(((.....)))))).......))).-- ( -22.60, z-score = -3.40, R) >droEre2.scaffold_4770 5771147 99 + 17746568 -UACAAAA-C-AAACAUCAUGG--AAAAUCUUCCUGCAUUACACAAACGCGGAA-AUUUUC-----------CUCG-UACU--CGCACAUUUCAUGGGUUUUCCACAGCUCAACCUGCA-- -.......-.-........(((--(((((((...............(((.(((.-....))-----------).))-)...--............))))))))))..((.......)).-- ( -17.81, z-score = -2.13, R) >droAna3.scaffold_13340 17983247 101 - 23697760 -CACAAAAGC-GAAAAUCAUGC--AAAAUCUUCCUGCAUUACACAAACGCGGAAGAUUUUC-----------ACCG-UACU--UGCUUGUUUCAUGGAUUUUUCAGAGCUCAACCUGCA-- -.....((((-((..((..((.--((((((((((.((...........)))))))))))))-----------)..)-)..)--)))))...................((.......)).-- ( -19.80, z-score = -0.82, R) >dp4.chr2 15651228 101 - 30794189 -GACAAAGGC-AAACAUCAUGC--AAAAUCUUCCUGCAUUACACAAACGCGGAAGGUUUUC-----------GCUG-UACU--CGCACAUUUCAUGGAUUUUCCAGCGCUCCAACUGCA-- -.......((-.........((--((((((((((.((...........)))))))))))).-----------))..-....--...........(((.....)))))((.......)).-- ( -20.50, z-score = -0.52, R) >droPer1.super_0 7061094 101 + 11822988 -GACAAAGGC-AAACAUCAUGC--AAAAUCUUCCUGCAUUACACAAACGCGGAAGGUUUUC-----------GCUG-UACU--CGCACAUUUCAUGGAUUUUCCAGCGCUCCAACUGCA-- -.......((-.........((--((((((((((.((...........)))))))))))).-----------))..-....--...........(((.....)))))((.......)).-- ( -20.50, z-score = -0.52, R) >droWil1.scaffold_181130 11123110 105 - 16660200 -CAAAAAG-C-AAAUAUCAUGC-AAAAAUCUUCCUGCAUUACACAAACGCGGAAGAUUUGC-----------CAUA-UACUUGCGCACAUUUCAUGAUUUUUCCAGUGCCCAACUACUACA -......(-(-((.(((.(((.-..(((((((((.((...........)))))))))))..-----------))))-)).))))((((.................))))............ ( -21.93, z-score = -3.04, R) >droVir3.scaffold_12822 1648891 110 - 4096053 -AAUAAAAGC-AAACAUCAUGC--AAAAUCUUCCUGCAUUACACAAACGCGGAAGAUUUUCUUGCUUUAAUACUUG-UACU--CGCACAUUUCAUGGAUUUUCCAGUGCUGA--CUGCA-- -....(((((-((.......(.--((((((((((.((...........))))))))))))))))))))......((-((.(--(((((......(((.....))))))).))--.))))-- ( -25.41, z-score = -2.71, R) >droMoj3.scaffold_6540 22685027 111 + 34148556 -AAUAAAAGCGAAAAAUCAUGC--AAAAUCUUCCUGCAUUACACAAACGCGGAAGAUUUGCUUGCUUUAAUACUUG-UACU--CGCACAUUUCAUGGAUUUCCCAAAGUUGA--CUGCA-- -....(((((((........((--(((.((((((.((...........))))))))))))))))))))........-....--.(((((.....(((.....)))....)).--.))).-- ( -21.90, z-score = -1.25, R) >droGri2.scaffold_14624 2694103 112 - 4233967 AUGCAAAA---CAACAUCGUGCAAAAAAUCUUCCUGAAUUACACAAACGCGGAAGAUUUUCUUGCUUUAAUGCUUGCUACU--CGCACAUUUCAUGGAUUUGCCAGUACUAA--CAGCA-- .(((....---((((((...((((((((((((((................)))))))))).))))....))).))).((((--.(((.(((.....))).))).))))....--..)))-- ( -23.99, z-score = -1.76, R) >consensus _UACAAAA_C_AAACAUCAUGC__AAAAUCUUCCUGCAUUACACAAACGCGGAAGAUUUUC___________CUCG_UACU__CGCACAUUUCAUGGAUUUUCCAGAGCUCAACCUGCA__ ........................((((((((((.((...........))))))))))))..................................(((.....)))................ (-11.55 = -11.76 + 0.21)
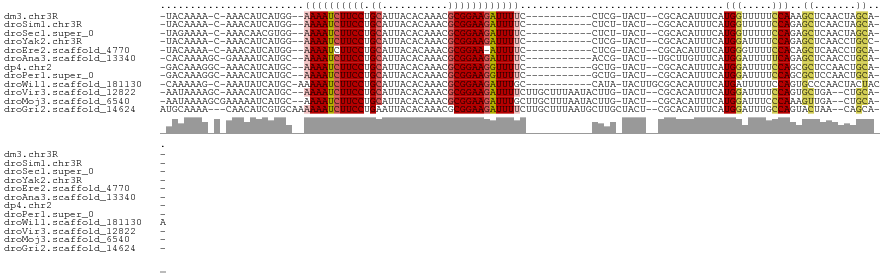
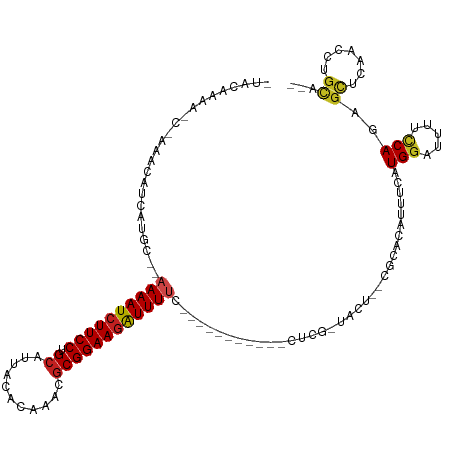
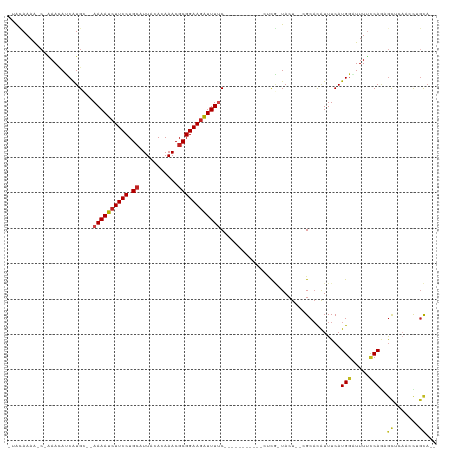
| Location | 9,656,756 – 9,656,860 |
|---|---|
| Length | 104 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 81.63 |
| Shannon entropy | 0.35496 |
| G+C content | 0.39819 |
| Mean single sequence MFE | -27.84 |
| Consensus MFE | -19.09 |
| Energy contribution | -19.94 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.47 |
| SVM RNA-class probability | 0.991256 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9656756 104 + 27905053 AGUACGAGGAAAAUCUUCCGCGUUUGUGUAAUGCAGGAAGAUUUUCCAUGAUGUUUG-UUUUGUAUUCGCUUUACAUUUUCCCCGAGCUGCUGAAAGUACAUAUG .((((..(((((((((((((((((.....))))).)))))))))))).....(((((-...((((.......)))).......)))))........))))..... ( -31.60, z-score = -3.97, R) >droSim1.chr3R 15746929 98 - 27517382 AGUAAGAGGAAAAUCUUCCGCGUUUGUGUAAUGCAGGAAGAUUUUCCAUGAUGUUUG-UUUUGUAUUCGCUUUACAUUUUCCCCAAGCGGCUGAAGAUA------ .......(((((((((((((((((.....))))).))))))))))))........((-((((.((..(((((............)))))..))))))))------ ( -28.90, z-score = -3.22, R) >droSec1.super_0 8769388 100 - 21120651 AGUAAGAGGAAAAUCUUCCGCGUUUGUGUAAUGCAGGAAGAUUUUCCACGUUGUUUG-UUUUCUAUACGCUUUACAUUUUCCCCAAGCGGCUGAAAAUAUA---- .......(((((((((((((((((.....))))).))))))))))))........((-(((((....(((((............)))))...)))))))..---- ( -32.50, z-score = -4.64, R) >droYak2.chr3R 13967093 100 + 28832112 AGUACGAGGAAAAUCUUCCGCGUUUGUGUAAUGCAGGAAGAUUUUCCAUGAUGUUUG-UUUUGUAUUCGCUUUUCAUUUUCCCCGAACUGCUGAAGACAUU---- .......(((((((((((((((((.....))))).))))))))))))..(((((((.-....(((((((..............)))).)))...)))))))---- ( -29.84, z-score = -3.65, R) >droEre2.scaffold_4770 5771183 99 - 17746568 AGUACGAGGAAAAU-UUCCGCGUUUGUGUAAUGCAGGAAGAUUUUCCAUGAUGUUUG-UUUUGUAUUCGCUUUAUAUACUCCCCGAACUGCUGAAGACAUU---- .......(((((((-(((((((((.....))))).)).)))))))))..(((((((.-....(((((((..............)))).)))...)))))))---- ( -22.54, z-score = -1.52, R) >droAna3.scaffold_13340 17983283 94 + 23697760 AGUACGGUGAAAAUCUUCCGCGUUUGUGUAAUGCAGGAAGAUUUUGCAUGAUUUUCGCUUUUGUGUUCGCCUUAAAUUUGCCACAAA-----AAAAGUU------ .....(((((((((((((((((((.....))))).))))))))).(((..(.........)..))))))))................-----.......------ ( -23.40, z-score = -1.62, R) >droPer1.super_0 7061130 96 - 11822988 AGUACAGCGAAAACCUUCCGCGUUUGUGUAAUGCAGGAAGAUUUUGCAUGAUGUUUGCCUUUGUCUUCACCU--CAUUCACUGCG-GCGGCAGCAGGUA------ ....((((((((..((((((((((.....))))).))))).)))))).)).....((((((((((....((.--((.....)).)-).))))).)))))------ ( -26.10, z-score = -0.09, R) >consensus AGUACGAGGAAAAUCUUCCGCGUUUGUGUAAUGCAGGAAGAUUUUCCAUGAUGUUUG_UUUUGUAUUCGCUUUACAUUUUCCCCGAGCGGCUGAAGAUA______ .......(((((((((((((((((.....))))).)))))))))))).......................................................... (-19.09 = -19.94 + 0.86)

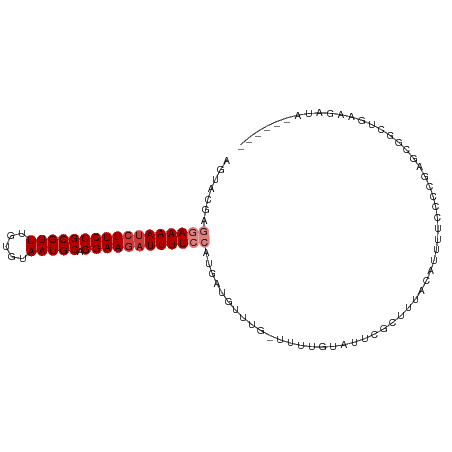
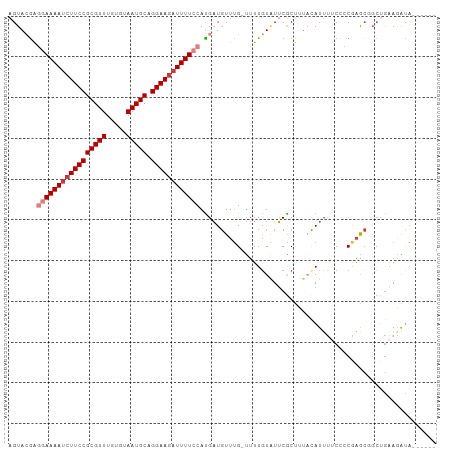
| Location | 9,656,756 – 9,656,860 |
|---|---|
| Length | 104 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 81.63 |
| Shannon entropy | 0.35496 |
| G+C content | 0.39819 |
| Mean single sequence MFE | -22.98 |
| Consensus MFE | -15.98 |
| Energy contribution | -16.03 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.970078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9656756 104 - 27905053 CAUAUGUACUUUCAGCAGCUCGGGGAAAAUGUAAAGCGAAUACAAAA-CAAACAUCAUGGAAAAUCUUCCUGCAUUACACAAACGCGGAAGAUUUUCCUCGUACU .....((((........(((..(........)..)))..........-..........((((((((((((.((...........))))))))))))))..)))). ( -23.30, z-score = -2.50, R) >droSim1.chr3R 15746929 98 + 27517382 ------UAUCUUCAGCCGCUUGGGGAAAAUGUAAAGCGAAUACAAAA-CAAACAUCAUGGAAAAUCUUCCUGCAUUACACAAACGCGGAAGAUUUUCCUCUUACU ------..(((((((....)))))))...((((.......))))...-..........((((((((((((.((...........))))))))))))))....... ( -23.70, z-score = -3.25, R) >droSec1.super_0 8769388 100 + 21120651 ----UAUAUUUUCAGCCGCUUGGGGAAAAUGUAAAGCGUAUAGAAAA-CAAACAACGUGGAAAAUCUUCCUGCAUUACACAAACGCGGAAGAUUUUCCUCUUACU ----(((((((((..((....)).)))))))))..((((........-......))))((((((((((((.((...........))))))))))))))....... ( -28.14, z-score = -3.97, R) >droYak2.chr3R 13967093 100 - 28832112 ----AAUGUCUUCAGCAGUUCGGGGAAAAUGAAAAGCGAAUACAAAA-CAAACAUCAUGGAAAAUCUUCCUGCAUUACACAAACGCGGAAGAUUUUCCUCGUACU ----..(((.(((.((..((((.......))))..))))).)))...-..........((((((((((((.((...........))))))))))))))....... ( -26.10, z-score = -3.42, R) >droEre2.scaffold_4770 5771183 99 + 17746568 ----AAUGUCUUCAGCAGUUCGGGGAGUAUAUAAAGCGAAUACAAAA-CAAACAUCAUGGAAAAUCUUCCUGCAUUACACAAACGCGGAA-AUUUUCCUCGUACU ----............(((.(((((((........(((.........-..........((((....))))((.....))....)))....-...))))))).))) ( -16.16, z-score = -0.42, R) >droAna3.scaffold_13340 17983283 94 - 23697760 ------AACUUUU-----UUUGUGGCAAAUUUAAGGCGAACACAAAAGCGAAAAUCAUGCAAAAUCUUCCUGCAUUACACAAACGCGGAAGAUUUUCACCGUACU ------.....((-----((((((................)))))))).........((.((((((((((.((...........))))))))))))))....... ( -18.09, z-score = -1.27, R) >droPer1.super_0 7061130 96 + 11822988 ------UACCUGCUGCCGC-CGCAGUGAAUG--AGGUGAAGACAAAGGCAAACAUCAUGCAAAAUCUUCCUGCAUUACACAAACGCGGAAGGUUUUCGCUGUACU ------.....(((((...-.))))).....--.(((((((((....(((.......))).....(((((.((...........))))))))))))))))..... ( -25.40, z-score = -0.77, R) >consensus ______UAUCUUCAGCAGCUCGGGGAAAAUGUAAAGCGAAUACAAAA_CAAACAUCAUGGAAAAUCUUCCUGCAUUACACAAACGCGGAAGAUUUUCCUCGUACU .........((((........)))).................................((((((((((((.((...........))))))))))))))....... (-15.98 = -16.03 + 0.05)

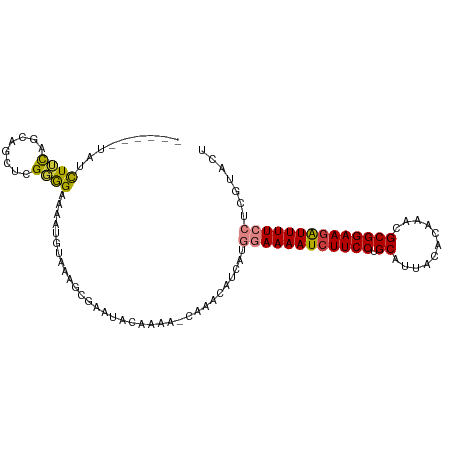
| Location | 9,656,764 – 9,656,876 |
|---|---|
| Length | 112 |
| Sequences | 8 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 68.73 |
| Shannon entropy | 0.60839 |
| G+C content | 0.38794 |
| Mean single sequence MFE | -25.87 |
| Consensus MFE | -13.74 |
| Energy contribution | -14.11 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.86 |
| SVM RNA-class probability | 0.995911 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9656764 112 + 27905053 ----GAAAAUCUUCCGCGUUUGUGUAAUGCAGGAAGAUUUU--CCAUGAUGUUUGU-UUUGUAUUCGCUUUACAUUUUCCCCGAGCUGCUGAAAGUACAUAUGUAUAUGCACGCUUGUA ----((((((((((((((((.....))))).))))))))))--)............-..((((.......)))).......((((((((.....((((....))))..))).))))).. ( -30.40, z-score = -3.07, R) >droSim1.chr3R 15746937 104 - 27517382 ----GAAAAUCUUCCGCGUUUGUGUAAUGCAGGAAGAUUUU--CCAUGAUGUUUGU-UUUGUAUUCGCUUUACAUUUUCCCCAAGCGGCUGAAG----AUAUAU----GCACGCUUGUC ----((((((((((((((((.....))))).))))))))))--)............-..((((.......)))).......((((((((.....----......----)).)))))).. ( -30.20, z-score = -3.25, R) >droSec1.super_0 8769396 108 - 21120651 ----GAAAAUCUUCCGCGUUUGUGUAAUGCAGGAAGAUUUU--CCACGUUGUUUGU-UUUCUAUACGCUUUACAUUUUCCCCAAGCGGCUGAAA----AUAUAUUUAUGCACGCUUGUC ----((((((((((((((((.....))))).))))))))))--)...((.((.(((-.....))).))...))........((((((((((((.----.....)))).)).)))))).. ( -30.70, z-score = -3.88, R) >droYak2.chr3R 13967101 108 + 28832112 ----GAAAAUCUUCCGCGUUUGUGUAAUGCAGGAAGAUUUU--CCAUGAUGUUUGU-UUUGUAUUCGCUUUUCAUUUUCCCCGAACUGCUGAAG----ACAUUUGUAUGCACGCUUGUA ----((((((((((((((((.....))))).))))))))))--).(..(.((.(((-..(((.(((((..(((.........)))..)).))).----))).......))).)))..). ( -26.20, z-score = -2.17, R) >droEre2.scaffold_4770 5771191 107 - 17746568 ----GAAAAU-UUCCGCGUUUGUGUAAUGCAGGAAGAUUUU--CCAUGAUGUUUGU-UUUGUAUUCGCUUUAUAUACUCCCCGAACUGCUGAAG----ACAUUUGUAUGCACUCUAGUA ----((((((-(((((((((.....))))).)).)))))))--)......(((((.-...((((.........))))....)))))(((((.((----.(((....)))..)).))))) ( -19.00, z-score = -0.21, R) >droAna3.scaffold_13340 17983291 99 + 23697760 ----GAAAAUCUUCCGCGUUUGUGUAAUGCAGGAAGAUUUU--GCAUGAUUUUCGCUUUUGUGUUCGCCUUAAAUUUGCCACAAA-AA---AAGUUUAGUACUCACAUG---------- ----.(((((((((((((((.....))))).))))))))))--.((((........(((((((...((.........))))))))-).---.(((.....)))..))))---------- ( -22.40, z-score = -1.63, R) >droPer1.super_0 7061138 88 - 11822988 ----GAAAACCUUCCGCGUUUGUGUAAUGCAGGAAGAUUUU--GCAUGAUGUUUGCCUUUGUCUUCACCUCAUUCACUGCGGCGGCAGCAGGUA------------------------- ----....((((.((((((..(((.((((..(((((((...--(((.......)))....)))))).)..))))))).)).))))....)))).------------------------- ( -24.90, z-score = -0.69, R) >droGri2.scaffold_14624 2694153 99 + 4233967 GCAAGAAAAUCUUCCGCGUUUGUGUAAUUCAGGAAGAUUUUUUGCACGAUGUUGUUUUGCAUUUGCCAUUAAAAAUAGACUUAUA-AAUGCAAGUUUACU------------------- ((((((((.((((((..(((.....)))...)))))))))))))).....((...((((((((((..................))-))))))))...)).------------------- ( -23.17, z-score = -1.95, R) >consensus ____GAAAAUCUUCCGCGUUUGUGUAAUGCAGGAAGAUUUU__CCAUGAUGUUUGU_UUUGUAUUCGCUUUACAUUUUCCCCAAACAGCUGAAG____AUAUUU_UAUGCACGCUUGU_ .....(((((((((((((((.....))))).)))))))))).............................................................................. (-13.74 = -14.11 + 0.37)
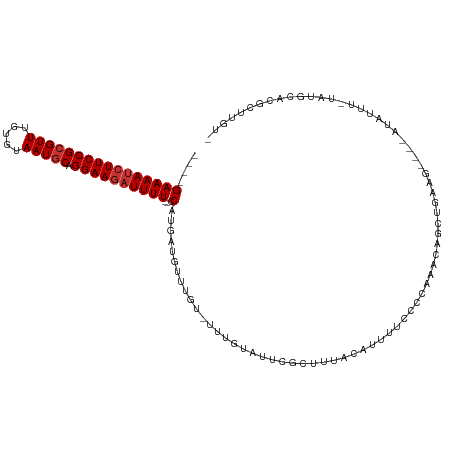
| Location | 9,656,764 – 9,656,876 |
|---|---|
| Length | 112 |
| Sequences | 8 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 68.73 |
| Shannon entropy | 0.60839 |
| G+C content | 0.38794 |
| Mean single sequence MFE | -21.82 |
| Consensus MFE | -10.05 |
| Energy contribution | -10.19 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.932728 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9656764 112 - 27905053 UACAAGCGUGCAUAUACAUAUGUACUUUCAGCAGCUCGGGGAAAAUGUAAAGCGAAUACAAA-ACAAACAUCAUGG--AAAAUCUUCCUGCAUUACACAAACGCGGAAGAUUUUC---- .....(((((((((....)))))))((((..(.....)..)))).......)).........-............(--((((((((((.((...........)))))))))))))---- ( -23.70, z-score = -2.21, R) >droSim1.chr3R 15746937 104 + 27517382 GACAAGCGUGC----AUAUAU----CUUCAGCCGCUUGGGGAAAAUGUAAAGCGAAUACAAA-ACAAACAUCAUGG--AAAAUCUUCCUGCAUUACACAAACGCGGAAGAUUUUC---- ..((((((.((----......----.....)))))))).......((((.......))))..-............(--((((((((((.((...........)))))))))))))---- ( -25.30, z-score = -2.67, R) >droSec1.super_0 8769396 108 + 21120651 GACAAGCGUGCAUAAAUAUAU----UUUCAGCCGCUUGGGGAAAAUGUAAAGCGUAUAGAAA-ACAAACAACGUGG--AAAAUCUUCCUGCAUUACACAAACGCGGAAGAUUUUC---- ..((((((.((..(((.....----)))..)))))))).............((((.......-.......)))).(--((((((((((.((...........)))))))))))))---- ( -26.04, z-score = -2.36, R) >droYak2.chr3R 13967101 108 - 28832112 UACAAGCGUGCAUACAAAUGU----CUUCAGCAGUUCGGGGAAAAUGAAAAGCGAAUACAAA-ACAAACAUCAUGG--AAAAUCUUCCUGCAUUACACAAACGCGGAAGAUUUUC---- ......((((........(((----.(((.((..((((.......))))..))))).)))..-........))))(--((((((((((.((...........)))))))))))))---- ( -22.57, z-score = -1.94, R) >droEre2.scaffold_4770 5771191 107 + 17746568 UACUAGAGUGCAUACAAAUGU----CUUCAGCAGUUCGGGGAGUAUAUAAAGCGAAUACAAA-ACAAACAUCAUGG--AAAAUCUUCCUGCAUUACACAAACGCGGAA-AUUUUC---- .....(((.((((....))))----.)))....(((((..............))))).....-............(--(((((.((((.((...........))))))-))))))---- ( -12.74, z-score = 1.11, R) >droAna3.scaffold_13340 17983291 99 - 23697760 ----------CAUGUGAGUACUAAACUU---UU-UUUGUGGCAAAUUUAAGGCGAACACAAAAGCGAAAAUCAUGC--AAAAUCUUCCUGCAUUACACAAACGCGGAAGAUUUUC---- ----------((((.((((.....))))---((-((((((................)))))))).......)))).--((((((((((.((...........)))))))))))).---- ( -19.09, z-score = -0.89, R) >droPer1.super_0 7061138 88 + 11822988 -------------------------UACCUGCUGCCGCCGCAGUGAAUGAGGUGAAGACAAAGGCAAACAUCAUGC--AAAAUCUUCCUGCAUUACACAAACGCGGAAGGUUUUC---- -------------------------.((((....((((....(((((((.((.(((((.....(((.......)))--....))))))).)))).)))....)))).))))....---- ( -25.10, z-score = -1.76, R) >droGri2.scaffold_14624 2694153 99 - 4233967 -------------------AGUAAACUUGCAUU-UAUAAGUCUAUUUUUAAUGGCAAAUGCAAAACAACAUCGUGCAAAAAAUCUUCCUGAAUUACACAAACGCGGAAGAUUUUCUUGC -------------------.......(((((((-(....(((..........)))))))))))...........((((((((((((((................)))))))))).)))) ( -19.99, z-score = -2.10, R) >consensus _ACAAGCGUGCAUA_AAAUAU____CUUCAGCAGCUUGGGGAAAAUGUAAAGCGAAUACAAA_ACAAACAUCAUGG__AAAAUCUUCCUGCAUUACACAAACGCGGAAGAUUUUC____ ..............................................................................((((((((((.((...........))))))))))))..... (-10.05 = -10.19 + 0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:13:20 2011