| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,370,568 – 7,370,658 |
| Length | 90 |
| Max. P | 0.940593 |

| Location | 7,370,568 – 7,370,658 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 69.68 |
| Shannon entropy | 0.57014 |
| G+C content | 0.56542 |
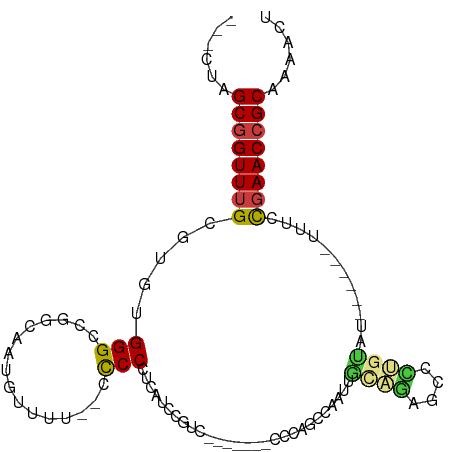
| Mean single sequence MFE | -24.71 |
| Consensus MFE | -12.29 |
| Energy contribution | -12.32 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.940593 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
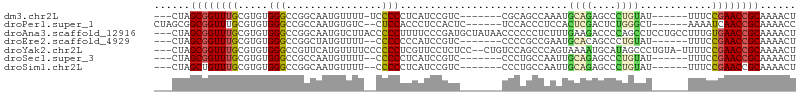
>dm3.chr2L 7370568 90 - 23011544 ---CUAGCGGUUUGCGUGUGGGCCGGCAAUGUUUU-UCCCCCUCAUCCGUC-------CGCAGCCAAAUGCAGAGCCCUGUAU------UUUCCGAACCGCAAAACU ---...(((((((((.(((((((.((..(((....-.......))))))))-------)))))).((((((((....))))))------))...)))))))...... ( -30.20, z-score = -2.64, R) >droPer1.super_1 8090824 93 + 10282868 CUAGCGGCGGUUUGCGUGUGGGCCGCCAAUGUGUC--CUCCACCCUCCACUC------UCCACCCUCCACUCGACUCUGGGCU------AAAAUCAACCGCAAAACC ...((((.((...(.(((..((.(((....))).)--)..))).).))....------....(((.............)))..------........))))...... ( -21.42, z-score = 0.06, R) >droAna3.scaffold_12916 15273981 104 - 16180835 ---CUAGCGGUUUGCGUGUGGGCCGGCAAUGUCUUACCCCCUUUUCCCGAUGCUAUAACCCCCCUCUUUGAAGACCCCAGCCUCCUGCCUUUGUGAACCGCAAAACU ---...((((((..((.(..((..(((...(((((...................................)))))....))).))..)...))..))))))...... ( -26.55, z-score = -1.99, R) >droEre2.scaffold_4929 16285469 89 - 26641161 ---CUAGCGGUUUGCGUGUGGGCCGGCUAUGUUUU--CCCCCCCAUCCGUC-------CCCCGCCGAAUGCACAGCCCUGUAU------UUUCCGAACCGCAAAACU ---...((((((((.(((.(((.(((..(((....--......)))))).)-------)).))).(((((((......)))))------))..))))))))...... ( -27.30, z-score = -2.26, R) >droYak2.chr2L 16791193 101 + 22324452 ---CUAGCGGUUUGCGUGUGGGCCGUUCAUGUUUUCCCCCCUCGUUCCUCUCC--CUGUCCAGCCCAGUAAAAUGCAUAGCCCUGUA-UUUUCCGAACCGCAAAACU ---...((((((((....(((((.(..((........................--.))..).)))))..((((((((......))))-)))).))))))))...... ( -24.81, z-score = -2.72, R) >droSec1.super_3 2885606 89 - 7220098 ---CUAGCGGUUUGCGUGUGGGCCGCCAAUGUUUU--CCCCCUCAUCCGUC-------CCCUGCCAAUUGCAGAGCCCUGUAU------UUUCCGAACCGCAAAACU ---...((((((((.(.(..((..((..(((....--......)))..)).-------.))..))...(((((....))))).------....))))))))...... ( -23.00, z-score = -1.56, R) >droSim1.chr2L 7158718 89 - 22036055 ---CUAGCUGUUUGCGUGUGGGCCGGCAAUGUUUU--CCCCCUCAUCCGUC-------CCCUGCCAAUUGCAGAGCCCUGUAU------UUUCCGAACCGCAAAACU ---...((.(((((.(((.(((.(((..(((....--......)))))).)-------)).)))....(((((....))))).------....))))).))...... ( -19.70, z-score = -0.34, R) >consensus ___CUAGCGGUUUGCGUGUGGGCCGGCAAUGUUUU__CCCCCUCAUCCGUC_______CCCAGCCAAUUGCAGAGCCCUGUAU______UUUCCGAACCGCAAAACU ......((((((((.....(((................)))............................((((....))))............))))))))...... (-12.29 = -12.32 + 0.02)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:23:29 2011