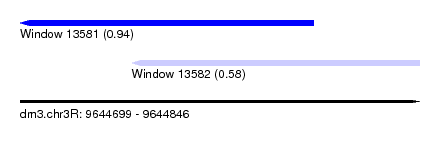
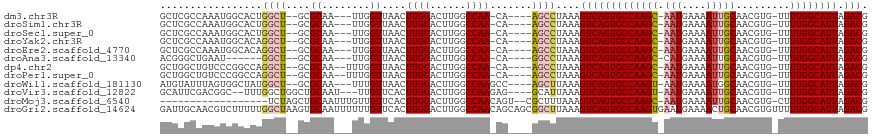
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,644,699 – 9,644,846 |
| Length | 147 |
| Max. P | 0.939142 |

| Location | 9,644,699 – 9,644,807 |
|---|---|
| Length | 108 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 78.23 |
| Shannon entropy | 0.44875 |
| G+C content | 0.46788 |
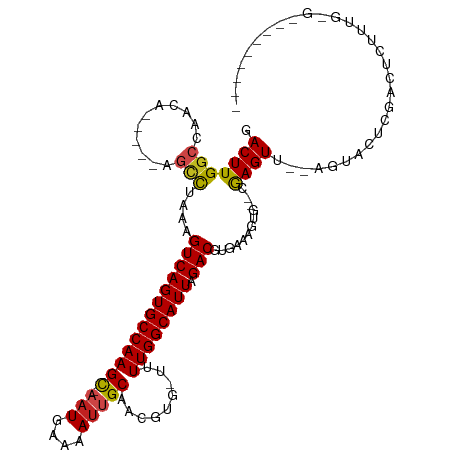
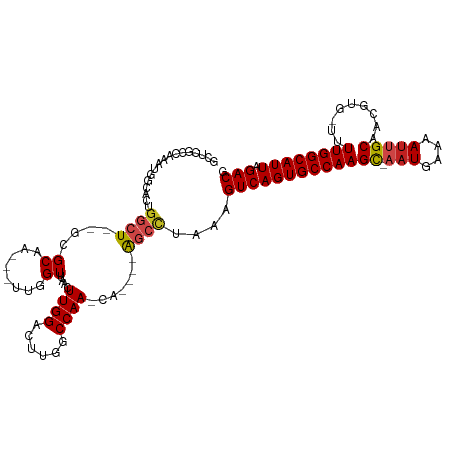
| Mean single sequence MFE | -34.53 |
| Consensus MFE | -16.96 |
| Energy contribution | -16.67 |
| Covariance contribution | -0.28 |
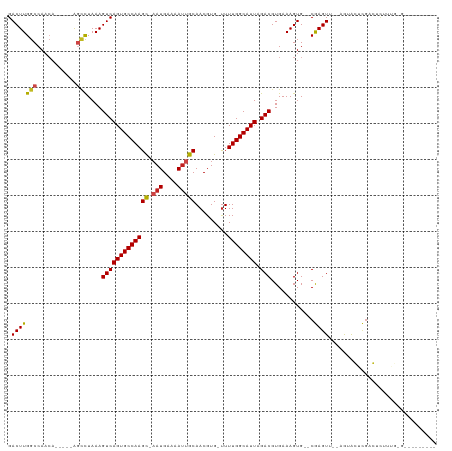
| Combinations/Pair | 1.14 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.939142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
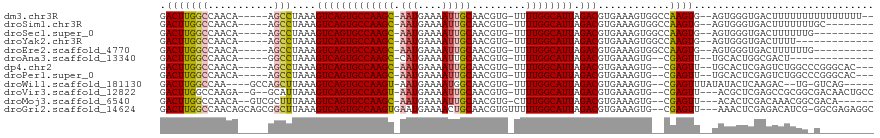
>dm3.chr3R 9644699 108 - 27905053 GACUUGGCCAACA-----AGCCUAAAGUCAGUGCCAAGC-AAUGAAAAUUGCAACGUG-UUUUGGCAUUAGACGUGAAAGUGGCCAAGUG--AGUGGGUGACUUUUUUUUUUUUUUU-- .(((((((((...-----.((.....(((((((((((((-(((....)))))......-..)))))))).))))).....)))))))))(--((.((....)).)))..........-- ( -34.60, z-score = -2.40, R) >droSim1.chr3R 15735048 102 + 27517382 GACUUGGCCAACA-----AGCCUAAAGUCAGUGCCAAGC-AAUGAAAAUUGCAACGUG-UUUUGGCAUUAGACGUGAAAGUGGCCAAGUG--AGUGGGUGACUUUUUUUGC-------- .(((((((((...-----.((.....(((((((((((((-(((....)))))......-..)))))))).))))).....)))))))))(--((.((....)).)))....-------- ( -34.60, z-score = -2.49, R) >droSec1.super_0 8757484 100 + 21120651 GACUUGGCCAACA-----AGCCUAAAGUCAGUGCCAAGC-AAUGAAAAUUGCAACGUG-UUUUGGCAUUAGACGUGAAAGUGGCCAAGUG--AGUGGGUGACUUUUUUG---------- .(((((((((...-----.((.....(((((((((((((-(((....)))))......-..)))))))).))))).....)))))))))(--((.((....)).)))..---------- ( -34.60, z-score = -2.82, R) >droYak2.chr3R 13954491 97 - 28832112 GACUUGGCCAACA-----AGCCUAAAGUCAGUGCCAAGC-AAUGAAAAUUGCAACGUG-UUUUGGCAUUAGACGUGAAAGUGGCCAAGUG--AGUGGGUGACUUUU------------- .(((((((((...-----.((.....(((((((((((((-(((....)))))......-..)))))))).))))).....))))))))).--...((....))...------------- ( -34.40, z-score = -3.02, R) >droEre2.scaffold_4770 5759240 100 + 17746568 GACUUGGCCAACA-----AGCCUAAAGUCAGUGCCAAGC-AAUGAAAAUUGCAACGUG-UUUUGGCAUUAGACGUGAAAGUGGCCAAGUG--AGUGGGUGACUUUUUUG---------- .(((((((((...-----.((.....(((((((((((((-(((....)))))......-..)))))))).))))).....)))))))))(--((.((....)).)))..---------- ( -34.60, z-score = -2.82, R) >droAna3.scaffold_13340 17969221 94 - 23697760 GACUUGGCCAACA-----GGCCUAAAGUCAGUGCCAAGC-CAUGAAAAUUGCAACGUG-UUUUGGCAUUAGACGUGAAAGUG--CGAGUU--UGCACUGGCGACU-------------- (((((((((....-----))))..)))))(((((((((.-((((..........))))-.)))))))))...(((...((((--((....--)))))).)))...-------------- ( -34.60, z-score = -2.76, R) >dp4.chr2 15639076 105 - 30794189 GACUUGGCCAACA-----AGCCUAAAGUCAGUGCCAAGC-AAUGAAAAUUGCAACGUG-UUUUGGCAUUAGACGUGAAAGUG--CGAGUU--UGCACUCGAGUCUGGCCCGGGCAC--- ((((((((.....-----.)))..))))).(((((..((-(((....)))))......-....(((..(((((.(((..(((--((....--)))))))).))))))))..)))))--- ( -38.40, z-score = -2.58, R) >droPer1.super_0 7048998 105 + 11822988 GACUUGGCCAACA-----AGCCUAAAGUCAGUGCCAAGC-AAUGAAAAUUGCAACGUG-UUUUGGCAUUAGACGUGAAAGUG--CGAGUU--UGCACUCGAGUCUGGCCCGGGCAC--- ((((((((.....-----.)))..))))).(((((..((-(((....)))))......-....(((..(((((.(((..(((--((....--)))))))).))))))))..)))))--- ( -38.40, z-score = -2.58, R) >droWil1.scaffold_181130 11101847 103 - 16660200 GACUUGGCCAA----GCCAGCUUAAAGUCAGUGCCAAGU-AAUGAAAAUGGCAACGUG-UUUUGGCAUUAGACGUGAAAGUG--CGAGUUUAUAUACUCAAGAC--UG-GUCAG----- ....((((((.----((..((((...((((((((((((.-.......(((....))).-.))))))))).)))....)))))--)((((......)))).....--))-)))).----- ( -31.40, z-score = -2.40, R) >droVir3.scaffold_12822 1632666 108 - 4096053 GACUUGGCCAAGA--G--GCAUUAAAGUCAGUGCCAAGU-AAUGAAAAUUGCAACGUG-UUUUGGCAUUAGACGUGAAAGUG--CGAGUU---ACGCUCGAGCCGCGGCGACAACUGCC ..((..(((((((--(--(((((......))))))..((-(((....)))))......-)))))))...))..(((...((.--((((..---...)))).)))))((((.....)))) ( -33.20, z-score = -1.03, R) >droMoj3.scaffold_6540 22670019 104 + 34148556 GACUUGGCCAACA--GUCGCUUUAAAGUCAGUGCCAAGC-AAUGAAAAUUGCAACGUG-CUUUGGCAUUAGACGUGAAAGUG--CGAGUU---ACACUCGACAAACGGCGACA------ ......(((....--..((((((...(((((((((((((-(((....)))))......-..)))))))).)))...))))))--((((..---...))))......)))....------ ( -34.40, z-score = -2.27, R) >droGri2.scaffold_14624 2680616 113 - 4233967 GACUUGGCCAACAGCAGCGGCUUAAAGUCAGUGCCAAGUGAAUGAAAACUGCAACGUGUUUUUGGCAUUAGACGUGAAAGUG--CGAGUU---AAACUCGAGACAUCG-GGCGAGAGGC ..((((.((.(((((....)))....((((((((((((...(((..........)))...))))))))).)))))....((.--((((..---...))))..))...)-).)))).... ( -31.20, z-score = -0.17, R) >consensus GACUUGGCCAACA_____AGCCUAAAGUCAGUGCCAAGC_AAUGAAAAUUGCAACGUG_UUUUGGCAUUAGACGUGAAAGUG__CGAGUU__AGUACUCGACUCUUUG_G_________ .(((((((...........)))....((((((((((((...(((..........)))...))))))))).)))............)))).............................. (-16.96 = -16.67 + -0.28)
| Location | 9,644,740 – 9,644,846 |
|---|---|
| Length | 106 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 81.75 |
| Shannon entropy | 0.39456 |
| G+C content | 0.47188 |
| Mean single sequence MFE | -36.01 |
| Consensus MFE | -17.74 |
| Energy contribution | -17.91 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.575256 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9644740 106 - 27905053 GCUCGCCAAAUGGCACUGGCU--GCGCAA---UUGGUUAACUUGGACUUGGCCAA-CA----AGCCUAAAGUCAGUGCCAAGC-AAUGAAAAUUGCAACGUG-UUUUGGCAUUAGACG ....(((((((((((((((((--..((..---((((((((.......))))))))-..----.))....))))))))))).((-(((....)))))......-.))))))........ ( -42.30, z-score = -3.78, R) >droSim1.chr3R 15735083 106 + 27517382 GCUCGCCAAAUGGCACUGGCU--GCGCAA---UUGGUUAACUUGGACUUGGCCAA-CA----AGCCUAAAGUCAGUGCCAAGC-AAUGAAAAUUGCAACGUG-UUUUGGCAUUAGACG ....(((((((((((((((((--..((..---((((((((.......))))))))-..----.))....))))))))))).((-(((....)))))......-.))))))........ ( -42.30, z-score = -3.78, R) >droSec1.super_0 8757517 106 + 21120651 GCUCGCCAAAUGGCACUGGCU--GCGCAA---UUGGUUAACUUGGACUUGGCCAA-CA----AGCCUAAAGUCAGUGCCAAGC-AAUGAAAAUUGCAACGUG-UUUUGGCAUUAGACG ....(((((((((((((((((--..((..---((((((((.......))))))))-..----.))....))))))))))).((-(((....)))))......-.))))))........ ( -42.30, z-score = -3.78, R) >droYak2.chr3R 13954521 106 - 28832112 GCUCGCCAAAUGGCACAGGCU--GCGCAA---UUGGUUAACUUGGACUUGGCCAA-CA----AGCCUAAAGUCAGUGCCAAGC-AAUGAAAAUUGCAACGUG-UUUUGGCAUUAGACG ....((((((((((((.((((--..((..---((((((((.......))))))))-..----.))....)))).)))))).((-(((....)))))......-.))))))........ ( -37.00, z-score = -2.14, R) >droEre2.scaffold_4770 5759273 106 + 17746568 GCUCGCCAAAUGGCACAGGCU--GCGCAA---UUGGUUAACUUGGACUUGGCCAA-CA----AGCCUAAAGUCAGUGCCAAGC-AAUGAAAAUUGCAACGUG-UUUUGGCAUUAGACG ....((((((((((((.((((--..((..---((((((((.......))))))))-..----.))....)))).)))))).((-(((....)))))......-.))))))........ ( -37.00, z-score = -2.14, R) >droAna3.scaffold_13340 17969248 100 - 23697760 ACGGGCUGAAU------GGCU--GCGCAA---UUGGUUAACUUGGACUUGGCCAA-CA----GGCCUAAAGUCAGUGCCAAGC-CAUGAAAAUUGCAACGUG-UUUUGGCAUUAGACG ..(((((...(------((((--(.....---..((((......)))))))))).-..----)))))...((((((((((((.-((((..........))))-.))))))))).))). ( -32.51, z-score = -1.19, R) >dp4.chr2 15639114 107 - 30794189 GCUGGCUGUCCCGGCCAGGCU--GCGCAA--UUUGGUUAACUUGGACUUGGCCAA-CA----AGCCUAAAGUCAGUGCCAAGC-AAUGAAAAUUGCAACGUG-UUUUGGCAUUAGACG .(((((((...)))))))(((--((((..--.(((((..(((..((((((((...-..----.)))..)))))))))))))((-(((....)))))...)))-)...)))........ ( -36.80, z-score = -1.20, R) >droPer1.super_0 7049036 107 + 11822988 GCUGGCUGUCCCGGCCAGGCU--GCGCAA--UUUGGUUAACUUGGACUUGGCCAA-CA----AGCCUAAAGUCAGUGCCAAGC-AAUGAAAAUUGCAACGUG-UUUUGGCAUUAGACG .(((((((...)))))))(((--((((..--.(((((..(((..((((((((...-..----.)))..)))))))))))))((-(((....)))))...)))-)...)))........ ( -36.80, z-score = -1.20, R) >droWil1.scaffold_181130 11101882 107 - 16660200 AUGUAUUUAGUGGCUAUGGCU--GCGCAA---UUUGUUAACUUGGACUUGGCCAAGCC----AGCUUAAAGUCAGUGCCAAGU-AAUGAAAAUGGCAACGUG-UUUUGGCAUUAGACG ..........(((((.(((((--.(.(((---.........))))....)))))))))----).......((((((((((((.-.......(((....))).-.))))))))).))). ( -29.10, z-score = -0.18, R) >droVir3.scaffold_12822 1632706 107 - 4096053 GCAUUCGACGGC--UUUGGCUGGCUGCAAU---UUGUUCACUUGGACUUGGCCAAGAG----GCAUUAAAGUCAGUGCCAAGU-AAUGAAAAUUGCAACGUG-UUUUGGCAUUAGACG (((.....((((--....))))..)))...---..((((....))))...((((((((----(((((......))))))..((-(((....)))))......-)))))))........ ( -30.90, z-score = -0.64, R) >droMoj3.scaffold_6540 22670053 96 + 34148556 ------------------UCUAGCUGCAAUUUGUUGUUCACUUGGACUUGGCCAACAGU--CGCUUUAAAGUCAGUGCCAAGC-AAUGAAAAUUGCAACGUG-CUUUGGCAUUAGACG ------------------.......((...((((((..((........))..)))))).--.))......(((((((((((((-(((....)))))......-..)))))))).))). ( -26.60, z-score = -0.81, R) >droGri2.scaffold_14624 2680655 118 - 4233967 GAUUGCAACGUCUUUUUGGCUAAGUGCAAUUUUUUGUUCACUUGGACUUGGCCAACAGCAGCGGCUUAAAGUCAGUGCCAAGUGAAUGAAAACUGCAACGUGUUUUUGGCAUUAGACG ........(((((.....((((((((((..((((..(((((((((..(((((....(((....)))....)))))..)))))))))..)))).)))).......))))))...))))) ( -38.51, z-score = -2.20, R) >consensus GCUCGCCAAAUGGCACUGGCU__GCGCAA___UUGGUUAACUUGGACUUGGCCAA_CA____AGCCUAAAGUCAGUGCCAAGC_AAUGAAAAUUGCAACGUG_UUUUGGCAUUAGACG .................((((.....(((............))).....)))).................((((((((((((...(((..........)))...))))))))).))). (-17.74 = -17.91 + 0.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:13:15 2011