| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,633,154 – 9,633,308 |
| Length | 154 |
| Max. P | 0.980699 |

| Location | 9,633,154 – 9,633,206 |
|---|---|
| Length | 52 |
| Sequences | 7 |
| Columns | 55 |
| Reading direction | reverse |
| Mean pairwise identity | 71.90 |
| Shannon entropy | 0.50685 |
| G+C content | 0.60423 |
| Mean single sequence MFE | -15.64 |
| Consensus MFE | -9.41 |
| Energy contribution | -9.99 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.980699 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3R 9633154 52 - 27905053 ---UUCCCCUGGCCGAUUCACCGGAUCCCCGGUGGAUCUCGCCGCCAUCUGCAAC ---.......(((.((((((((((....))))))))))..)))((.....))... ( -22.60, z-score = -3.88, R) >droSim1.chr3R 15723420 52 + 27517382 ---UUCCCCUGGCCGAUUCACCGGAUCCCCGGUGGAUCUCGCCGCCAUCUGCAAC ---.......(((.((((((((((....))))))))))..)))((.....))... ( -22.60, z-score = -3.88, R) >droSec1.super_0 8745981 52 + 21120651 ---UUCCCCCGGCCGAUUCACCGGAUCCCCGGUGGAUCUCGCCGCCAUCUGCAAC ---......((((.((((((((((....))))))))))..))))........... ( -23.40, z-score = -4.00, R) >droYak2.chr3R 13942469 52 - 28832112 ---CUCCCCUACCCGAUUCUCCGGAUCCCCGGUGGAUCUUGCCGCCAUCUGCAAC ---...........(((((.((((....)))).)))))((((........)))). ( -14.70, z-score = -2.38, R) >droEre2.scaffold_4770 5747344 52 + 17746568 ---UUCCCCCGCCCGAUUCCCCGGAUCCUCGGUGGAUCUCGCCGCCAUCUGCAAC ---.......(((((......)))......(((((......)))))....))... ( -13.20, z-score = -0.67, R) >dp4.chr2 15628179 51 - 30794189 GAUUUCUCUGCCCCCAACCAUUUGAUGUUUGGUG--UUUGGU--UCGACUGCAAC ....((...(((....((((.........)))).--...)))--..))....... ( -6.50, z-score = 0.75, R) >droPer1.super_0 7038158 51 + 11822988 GAUUUCUCUGCCCCCAACCAUUUGAUGUUUGGUG--UUUGGU--UCGACUGCAAC ....((...(((....((((.........)))).--...)))--..))....... ( -6.50, z-score = 0.75, R) >consensus ___UUCCCCUGCCCGAUUCACCGGAUCCCCGGUGGAUCUCGCCGCCAUCUGCAAC ..............((((((((((....))))))))))..((........))... ( -9.41 = -9.99 + 0.58)
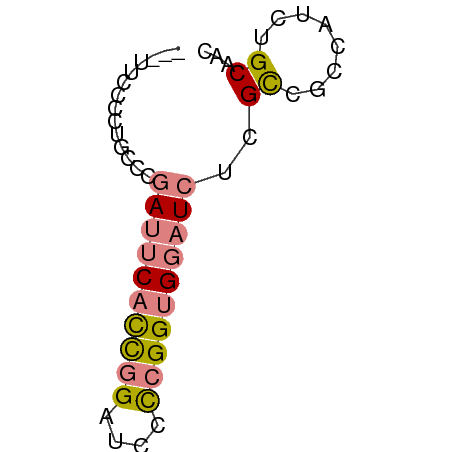
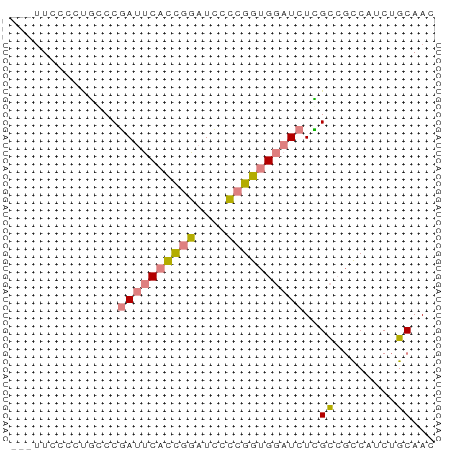
| Location | 9,633,206 – 9,633,308 |
|---|---|
| Length | 102 |
| Sequences | 11 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 75.31 |
| Shannon entropy | 0.51606 |
| G+C content | 0.48572 |
| Mean single sequence MFE | -18.80 |
| Consensus MFE | -11.10 |
| Energy contribution | -11.47 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.776556 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9633206 102 + 27905053 AUCACAUUUCUCUGCGAGUUCAUUCAACCUCGCCCCAGUGAACUUAGCUGAGGUUAUUUUUACUUCCAAGCAACAGCCUCCU---CCGGAAUCCCCCAGCUCCGA---------- .............(((((((((((............))))))))).((((((((.......)))))..)))....)).....---.((((..........)))).---------- ( -18.20, z-score = -0.77, R) >droPer1.super_0 7038209 96 - 11822988 AUCACAUUUCUCUGGGAGUUCAUUCAACCUCGCCCCAGUGAACUUAGCUGAGGUUAUUUUUACUUCCAAGCAACA-UUGCCU---CUCCUCUCGCUCUGC--------------- ............(((((((......((((((((...((....))..)).))))))......))))))).(((...-.)))..---...............--------------- ( -20.50, z-score = -1.57, R) >dp4.chr2 15628230 96 + 30794189 AUCACAUUUCUCUGGGAGUUCAUUCAACCUCGCCCCAGUGAACUUAGCUGAGGUUAUUUUUACUUCCAAGCAACA-UUGCCU---CUCCUCUCGCUCUGC--------------- ............(((((((......((((((((...((....))..)).))))))......))))))).(((...-.)))..---...............--------------- ( -20.50, z-score = -1.57, R) >droAna3.scaffold_13340 17958119 81 + 23697760 AUCACAUUUCUCUACGAGUUCAUUCAACCUCGCCCCAGUGAACUUAACUGAGGUUAUUUUUACUCCCAAGAAAC-----UCC---CCGG-------------------------- ......(((((....((((......(((((((....((....))....)))))))......))))...))))).-----...---....-------------------------- ( -15.00, z-score = -2.33, R) >droEre2.scaffold_4770 5747396 100 - 17746568 AUCACAUUUCUCUGCGAGUUCAUUCAACCUCGCCCCAGUGAACUUAGCUGAGGUUAUUUUUACUUCCAAGCAACAGCCUCCU---CCGGAAUCCCCCGACUCU------------ .............(((((((((((............))))))))).((((((((.......)))))..)))....)).....---.(((......))).....------------ ( -17.90, z-score = -1.11, R) >droYak2.chr3R 13942521 100 + 28832112 AUCACAUUUCUCUGCGAGUUCAUUCAACCUCGCCCCAGUGAACUUAGCUGAGGUUAUUUUUACUUCCAAGCAACAGCCUCCU---CCGGAAUCCCCCGACUCC------------ .............(((((((((((............))))))))).((((((((.......)))))..)))....)).....---.(((......))).....------------ ( -17.90, z-score = -1.16, R) >droSec1.super_0 8746033 102 - 21120651 AUCACAUUUCUCUGCGAGUUCAUUCAACCUCGCCCCAGUGAACUUAGCUGAGGUUAUUUUUACUUCCAAGCAACAGCCUCCU---CCGGAAUCCCCCAGCUCCAA---------- .((((........(((((..........)))))....))))....(((((.((..(((((.........((....)).....---..)))))..)))))))....---------- ( -17.49, z-score = -1.02, R) >droSim1.chr3R 15723472 102 - 27517382 AUCACAUUUCUCUGCGAGUUCAUUCAACCUCGCCCCAGUGAACUUAGCUGAGGUUAUUUUUACUUCCAAGCAACAGCCUCCU---CCGGAAUCCCCCAGCUCCAA---------- .((((........(((((..........)))))....))))....(((((.((..(((((.........((....)).....---..)))))..)))))))....---------- ( -17.49, z-score = -1.02, R) >droVir3.scaffold_12822 1619548 102 + 4096053 ---AUAUUGCUGUGCAAGUUCAUUCAACCUCGCCCCAGUGAACUUAGCUAAGGUUAUUUUUACUUCCAAGCUAGAAAUGUUU--GCCGCCCGUGCCGCAUACUCCCC-------- ---....(((.(..((((((((((............)))))))))......(((((((((((((....)).))))))))...--)))....)..).)))........-------- ( -20.00, z-score = -1.20, R) >droMoj3.scaffold_6540 22653764 112 - 34148556 AGCACAUUGCUGAGGGAGUUCAUUCAACCUCGCCCCAGUGAACUUAGCUGAGGUUAUUUUUACAUCCAAGCUAGAAAUUUU---GCCCCCUUGCCCUUGCAACCACCCGCUCCAC .(((....((.(((((.((......((((((((...((....))..)).))))))((((((((......).)))))))...---)).)))))))...)))............... ( -23.10, z-score = -0.45, R) >droGri2.scaffold_14624 2666450 100 + 4233967 ---------------GAGUUCAUUCAACCUCGCCCCAGUGAACUUAGCUGAGGUUAUUUUUACAUCCAAGCUAGAAAUUUGCUGGCCCCCCUCCCCUCCCUCCCAGCUGGUCUAC ---------------..((......((((((((...((....))..)).))))))......))........((((.....(((((.................)))))...)))). ( -18.73, z-score = -0.40, R) >consensus AUCACAUUUCUCUGCGAGUUCAUUCAACCUCGCCCCAGUGAACUUAGCUGAGGUUAUUUUUACUUCCAAGCAACAGCCUCCU___CCGGAAUCCCCCAGCUCC____________ ...............(((((((((............))))))))).((((((((.......)))))..)))............................................ (-11.10 = -11.47 + 0.37)

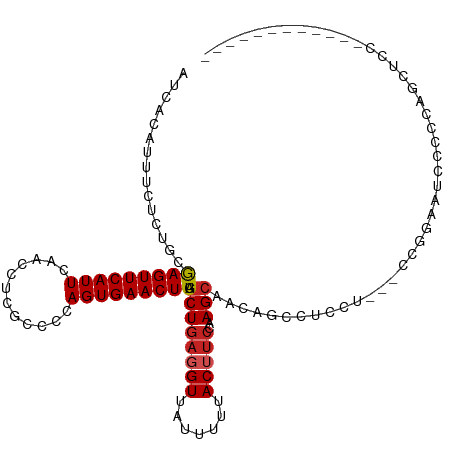
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:13:13 2011