| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,616,420 – 9,616,498 |
| Length | 78 |
| Max. P | 0.797928 |

| Location | 9,616,420 – 9,616,498 |
|---|---|
| Length | 78 |
| Sequences | 11 |
| Columns | 82 |
| Reading direction | forward |
| Mean pairwise identity | 90.53 |
| Shannon entropy | 0.20495 |
| G+C content | 0.35343 |
| Mean single sequence MFE | -18.05 |
| Consensus MFE | -11.64 |
| Energy contribution | -12.51 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.719575 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
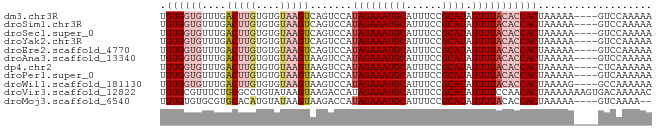
>dm3.chr3R 9616420 78 + 27905053 UGUGGUGUUUGACUUGUGUGUAAGUCAGUCCAUAGAAAUGCAUUUCCGCAUAUUUUACACCACUAAAAA----GUCCAAAAA .((((((.((((((((....))))))))....(((((((((......)))).)))))))))))......----......... ( -20.20, z-score = -3.24, R) >droSim1.chr3R 15704831 78 - 27517382 UGUGGUGUUUGACUUGUGUGUAAGUCAGUCCAUAGAAAUGCAUUUCCGCAUAUUUUACACCACUAAAAA----GUCCAAAAA .((((((.((((((((....))))))))....(((((((((......)))).)))))))))))......----......... ( -20.20, z-score = -3.24, R) >droSec1.super_0 8729643 78 - 21120651 UGUGGUGUUUGACUUGUGUGUAAGUCAGUCCAUAGAAAUGCAUUUCCGCAUAUUUUACACCACUAAAAA----GUCCAAAAA .((((((.((((((((....))))))))....(((((((((......)))).)))))))))))......----......... ( -20.20, z-score = -3.24, R) >droYak2.chr3R 13925376 78 + 28832112 UGUGGUGUUUGACUUGUGUGUAAGUCAGUCCAUAGAAAUGCAUUUCCGCAUAUUUUACACCACUAAAAA----GUCCAAAAA .((((((.((((((((....))))))))....(((((((((......)))).)))))))))))......----......... ( -20.20, z-score = -3.24, R) >droEre2.scaffold_4770 5730562 78 - 17746568 UGUGGUGUUUGACUUGUGUGUAAGUCAGUCCAUAGAAAUGCAUUUCCGCAUAUUUUACACCACUAAAAA----GUCCAAAAA .((((((.((((((((....))))))))....(((((((((......)))).)))))))))))......----......... ( -20.20, z-score = -3.24, R) >droAna3.scaffold_13340 17941634 78 + 23697760 UGUGGUGUUUGACUUGUGUGUAAGUAAGUCCAUAGAAAUGCAUUUCCGCAUAUUUUACACCACUAAAAA----GUCCAAAAA .((((((...((((((........))))))..(((((((((......)))).)))))))))))......----......... ( -18.00, z-score = -2.54, R) >dp4.chr2 15612107 78 + 30794189 UGUGGUGUUUGACUUGUGUGUAAGUAAGUCCAUAGAAAUGCAUUUCCGCAUAUUUUACACCACUAAAAA----CUCAAAAAA .((((((...((((((........))))))..(((((((((......)))).)))))))))))......----......... ( -18.00, z-score = -2.45, R) >droPer1.super_0 7021948 78 - 11822988 UGUGGUGUUUGACUUGUGUGUAAGUAAGUCCAUAGAAAUGCAUUUCCGCAUAUUUUACACCACUAAAAA----GUCAAAAAA .((((((...((((((........))))))..(((((((((......)))).)))))))))))......----......... ( -18.00, z-score = -2.48, R) >droWil1.scaffold_181130 11070568 78 + 16660200 UGUGGUGUUUGACUUGUGUGUAAGUAAGUCCAUAGAAAUGCAUUUCCGCAUAUUUUACACCACUAAAAG----GCCAAAAAA .((((((...((((((........))))))..(((((((((......)))).)))))))))))......----......... ( -18.00, z-score = -1.73, R) >droVir3.scaffold_12822 1601439 82 + 4096053 UGUGCGUUUCUGCGCCUGUAUAAGUAAGACCAUAGAAAUGCAUUUCCGCAUAUUUUCCAACACUAAAAAAAGUGACAAAAAC .(((((((((((.(.((.........))..).))))))))))).................((((......))))........ ( -13.70, z-score = -1.17, R) >droMoj3.scaffold_6540 22633465 76 - 34148556 UGUGUGUGCGUGCACAUGUAUAAGUAAGACCAUAGAAAUGCAUUUCCGCAUAUUUUACACCACUAAAAA----GUCAAAA-- ((..((((....))))..)).......(((.......((((......)))).(((((......))))).----)))....-- ( -11.90, z-score = 0.03, R) >consensus UGUGGUGUUUGACUUGUGUGUAAGUAAGUCCAUAGAAAUGCAUUUCCGCAUAUUUUACACCACUAAAAA____GUCCAAAAA .((((((....(((((....))))).......(((((((((......)))).)))))))))))................... (-11.64 = -12.51 + 0.87)


| Location | 9,616,420 – 9,616,498 |
|---|---|
| Length | 78 |
| Sequences | 11 |
| Columns | 82 |
| Reading direction | reverse |
| Mean pairwise identity | 90.53 |
| Shannon entropy | 0.20495 |
| G+C content | 0.35343 |
| Mean single sequence MFE | -18.35 |
| Consensus MFE | -13.06 |
| Energy contribution | -14.09 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.797928 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
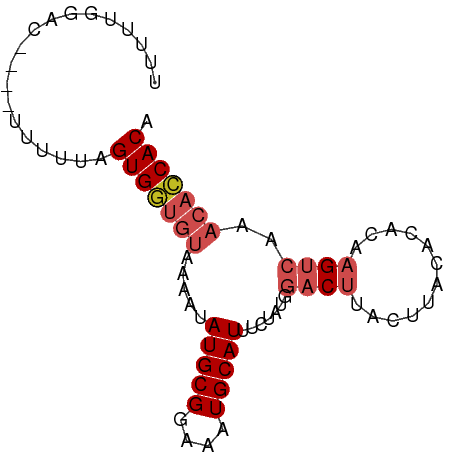
>dm3.chr3R 9616420 78 - 27905053 UUUUUGGAC----UUUUUAGUGGUGUAAAAUAUGCGGAAAUGCAUUUCUAUGGACUGACUUACACACAAGUCAAACACCACA .........----......(((((((.....(((((....)))))..........((((((......)))))).))))))). ( -20.00, z-score = -2.90, R) >droSim1.chr3R 15704831 78 + 27517382 UUUUUGGAC----UUUUUAGUGGUGUAAAAUAUGCGGAAAUGCAUUUCUAUGGACUGACUUACACACAAGUCAAACACCACA .........----......(((((((.....(((((....)))))..........((((((......)))))).))))))). ( -20.00, z-score = -2.90, R) >droSec1.super_0 8729643 78 + 21120651 UUUUUGGAC----UUUUUAGUGGUGUAAAAUAUGCGGAAAUGCAUUUCUAUGGACUGACUUACACACAAGUCAAACACCACA .........----......(((((((.....(((((....)))))..........((((((......)))))).))))))). ( -20.00, z-score = -2.90, R) >droYak2.chr3R 13925376 78 - 28832112 UUUUUGGAC----UUUUUAGUGGUGUAAAAUAUGCGGAAAUGCAUUUCUAUGGACUGACUUACACACAAGUCAAACACCACA .........----......(((((((.....(((((....)))))..........((((((......)))))).))))))). ( -20.00, z-score = -2.90, R) >droEre2.scaffold_4770 5730562 78 + 17746568 UUUUUGGAC----UUUUUAGUGGUGUAAAAUAUGCGGAAAUGCAUUUCUAUGGACUGACUUACACACAAGUCAAACACCACA .........----......(((((((.....(((((....)))))..........((((((......)))))).))))))). ( -20.00, z-score = -2.90, R) >droAna3.scaffold_13340 17941634 78 - 23697760 UUUUUGGAC----UUUUUAGUGGUGUAAAAUAUGCGGAAAUGCAUUUCUAUGGACUUACUUACACACAAGUCAAACACCACA .........----......(((((((.....(((((....))))).......(((((..........)))))..))))))). ( -18.10, z-score = -2.54, R) >dp4.chr2 15612107 78 - 30794189 UUUUUUGAG----UUUUUAGUGGUGUAAAAUAUGCGGAAAUGCAUUUCUAUGGACUUACUUACACACAAGUCAAACACCACA .........----......(((((((.....(((((....))))).......(((((..........)))))..))))))). ( -18.10, z-score = -2.23, R) >droPer1.super_0 7021948 78 + 11822988 UUUUUUGAC----UUUUUAGUGGUGUAAAAUAUGCGGAAAUGCAUUUCUAUGGACUUACUUACACACAAGUCAAACACCACA .........----......(((((((.....(((((....))))).......(((((..........)))))..))))))). ( -18.10, z-score = -2.78, R) >droWil1.scaffold_181130 11070568 78 - 16660200 UUUUUUGGC----CUUUUAGUGGUGUAAAAUAUGCGGAAAUGCAUUUCUAUGGACUUACUUACACACAAGUCAAACACCACA .........----......(((((((.....(((((....))))).......(((((..........)))))..))))))). ( -18.10, z-score = -2.11, R) >droVir3.scaffold_12822 1601439 82 - 4096053 GUUUUUGUCACUUUUUUUAGUGUUGGAAAAUAUGCGGAAAUGCAUUUCUAUGGUCUUACUUAUACAGGCGCAGAAACGCACA ((((((((((((......)))).(((((...(((((....))))))))))..((((.........))))))))))))..... ( -16.90, z-score = -0.87, R) >droMoj3.scaffold_6540 22633465 76 + 34148556 --UUUUGAC----UUUUUAGUGGUGUAAAAUAUGCGGAAAUGCAUUUCUAUGGUCUUACUUAUACAUGUGCACGCACACACA --.......----......((((((((....(((((....)))))...((((............))))))))).)))..... ( -12.60, z-score = 0.30, R) >consensus UUUUUGGAC____UUUUUAGUGGUGUAAAAUAUGCGGAAAUGCAUUUCUAUGGACUUACUUACACACAAGUCAAACACCACA ...................(((((((.....(((((....))))).......((((............))))..))))))). (-13.06 = -14.09 + 1.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:13:11 2011