
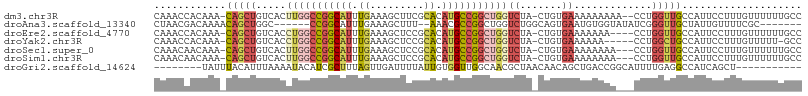
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,607,981 – 9,608,085 |
| Length | 104 |
| Max. P | 0.524378 |

| Location | 9,607,981 – 9,608,085 |
|---|---|
| Length | 104 |
| Sequences | 7 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 67.63 |
| Shannon entropy | 0.65342 |
| G+C content | 0.47418 |
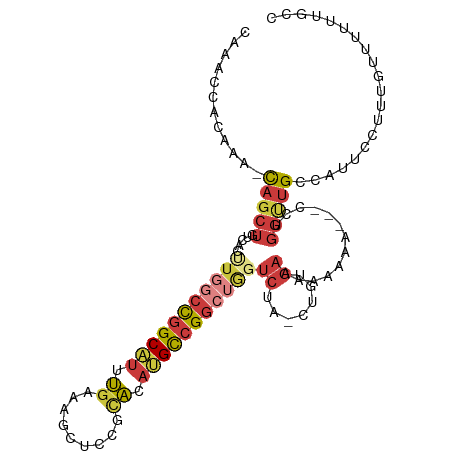
| Mean single sequence MFE | -25.92 |
| Consensus MFE | -12.39 |
| Energy contribution | -13.73 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.45 |
| Mean z-score | -0.66 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.524378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9607981 104 + 27905053 CAAACCACAAA-CAGCUGUCACUUGGCCGGCAUUUGAAAGCUUCGCACAUGCCGGCUGGUCUA-CUGUGAAAAAAAAA--CCUGGUUGCCAUUCCUUUGUUUUUUGCC ..........(-(((..(....(..((((((((.((.........)).))))))))..).)..-))))....((((((--(..((........))...)))))))... ( -25.90, z-score = -0.28, R) >droAna3.scaffold_13340 17933473 93 + 23697760 CUAACGACAAAACAGCUGGC------CCGGCAUUUGAAAGCUUU--AAACGCCGGCUGGUCUGGCAGUGAAUGUGGUAUAUCGGGUUGCUAUUGUUUUCGC------- ............((((..((------(.(((.(((((.....))--))).))))))..).)))...(((((.((((((........))))))....)))))------- ( -27.10, z-score = -0.53, R) >droEre2.scaffold_4770 5722057 102 - 17746568 CAAACCACAAA-CAGCUGUCACCUGGCCGGCAUUUGAAAGCUCCGCACAUGCCGGCUGGUCUA-CUGUGAAAAAAA----CCUGGUUGCCAUUCCUUUGUUUUUUGCC ..........(-(((..(..(((.(((((((((.((.........)).)))))))))))))..-))))..((((((----(..((........))...)))))))... ( -27.60, z-score = -0.72, R) >droYak2.chr3R 13916772 100 + 28832112 CAAACCACAAA-CAGCUGUCACCUGGCCGGCAUUUGAAAGCUCCGCACAUGCCGGCUGGUCUA-CUGUGAAAAAA-----CCUGGCUGCCAUUCCUUUGUUUUU-GCC ((((..(((((-(((((.(((((..((((((((.((.........)).))))))))..)....-..)))).....-----...))))).......))))).)))-).. ( -26.41, z-score = -0.32, R) >droSec1.super_0 8721230 103 - 21120651 CAAACAACAAA-CAGCUGUCACUUGGCCGGCAUUUGAAAGCUCCGCACAUGCCGGCUGGUCUA-CUGUGAAAAAAAA---CCUGGUUGCCAUUCCUUUGUUUUUUGCC ((((.((((((-(((..(....(..((((((((.((.........)).))))))))..).)..-)))).........---...((........)).))))).)))).. ( -26.10, z-score = -0.38, R) >droSim1.chr3R 15696335 103 - 27517382 CAAACAACAAA-CAGCUGUCACUUGGCCGGCAUUUGAAAGCUCCGCACAUGCCGGCUGGUCUA-CUGUGAAAAAAAA---CCUGGUUGCCAUUCCUUUGUUUUUUGCC ((((.((((((-(((..(....(..((((((((.((.........)).))))))))..).)..-)))).........---...((........)).))))).)))).. ( -26.10, z-score = -0.38, R) >droGri2.scaffold_14624 2631306 89 + 4233967 --------UAUUUACAUUUAAAAUACAUCGCUUUAGUUGAUUUUAUUGUGGUUGGCAACGCUAACAACAGCUGACCGGCAUUUUGAGGCCAUCAGCU----------- --------....((((..((((((.((.(......).)))))))).))))((((((...))))))...((((((..(((........))).))))))----------- ( -22.20, z-score = -2.04, R) >consensus CAAACCACAAA_CAGCUGUCACUUGGCCGGCAUUUGAAAGCUCCGCACAUGCCGGCUGGUCUA_CUGUGAAAAAAAA___CCUGGUUGCCAUUCCUUUGUUUUUUGCC ............(((((.....(((((((((((.((.........)).)))))))))))((.......)).............))))).................... (-12.39 = -13.73 + 1.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:13:09 2011