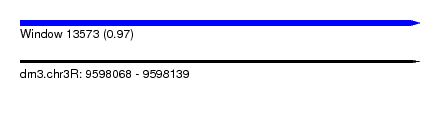
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,598,068 – 9,598,139 |
| Length | 71 |
| Max. P | 0.971796 |

| Location | 9,598,068 – 9,598,139 |
|---|---|
| Length | 71 |
| Sequences | 13 |
| Columns | 76 |
| Reading direction | forward |
| Mean pairwise identity | 85.49 |
| Shannon entropy | 0.33459 |
| G+C content | 0.47277 |
| Mean single sequence MFE | -22.85 |
| Consensus MFE | -19.51 |
| Energy contribution | -19.46 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.971796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
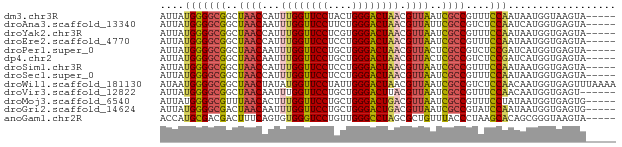
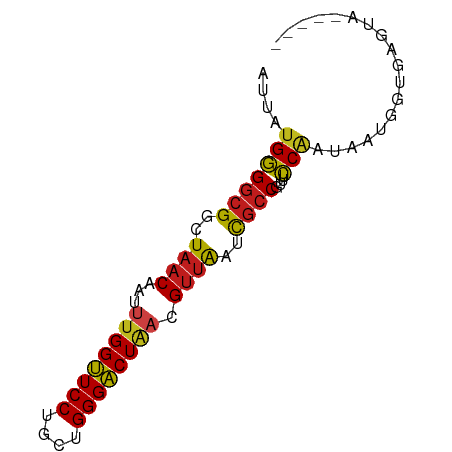
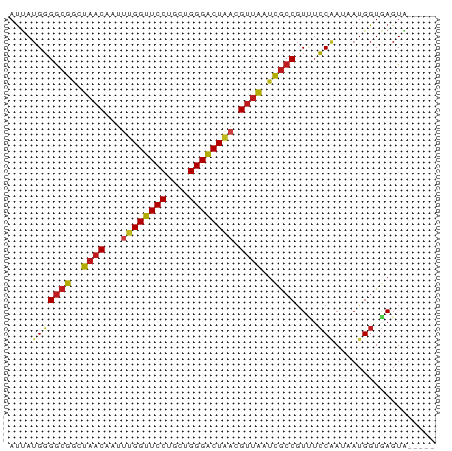
>dm3.chr3R 9598068 71 + 27905053 AUUAUGGGGCGGCUAACCAUUUGGUUCCUACUGGGACUAACGUUAAUCGCCGUUUCCAAUAAUGGUAAGUA----- ((((((..(((((((((...((((((((....)))))))).))))...)))))..)..)))))........----- ( -22.40, z-score = -2.11, R) >droAna3.scaffold_13340 17923683 71 + 23697760 AUUAUGGGGCGGCUAACAAUUUGGUUCCUUCUGGGACUAACGUUAUUCGCCGUCUCCAAUCAUGGUGAGUA----- ....(((((((((((((...((((((((....)))))))).))))...)))))).))).............----- ( -24.60, z-score = -3.05, R) >droYak2.chr3R 13906629 71 + 28832112 AUUAUGGGGCGGCUAACCAUUUGGUUCCUCGUGGGACUAACGUUAAUCGCCGUUUCCAAUAAUGGUGAGUA----- ((((((..(((((((((...((((((((....)))))))).))))...)))))..)..)))))........----- ( -22.40, z-score = -1.68, R) >droEre2.scaffold_4770 5711743 71 - 17746568 AUUAUGGGGCGGCUAACCAUUUGGUUCCUCCUGGGACUAACGUUAAUCGCCGUUUCCAAUAAUGGUGAGUA----- ((((((..(((((((((...((((((((....)))))))).))))...)))))..)..)))))........----- ( -22.40, z-score = -1.64, R) >droPer1.super_0 1488288 71 - 11822988 AUUAUGGGGCGGCUAACAAUUUGGUUCCUGCUGGGACUAACGUUACUCGCCGUCUCCGAUCAUGGUGAGUA----- ....(((((((((((((...((((((((....)))))))).))))...)))))).))).............----- ( -23.90, z-score = -1.92, R) >dp4.chr2 10146617 71 + 30794189 AUUAUGGGGCGGCUAACAAUUUGGUUCCUGCUGGGACUAACGUUACUCGCCGUCUCCGAUCAUGGUGAGUA----- ....(((((((((((((...((((((((....)))))))).))))...)))))).))).............----- ( -23.90, z-score = -1.92, R) >droSim1.chr3R 15688969 71 - 27517382 AUUAUGGGGCGGCUAACCAUUUGGUUCCUCCUGGGACUAACGUUAAUCGCCGUUUCCAAUAAUGGUGAGUA----- ((((((..(((((((((...((((((((....)))))))).))))...)))))..)..)))))........----- ( -22.40, z-score = -1.64, R) >droSec1.super_0 8711321 71 - 21120651 AUUAUGGGGCGGCUAACCAUUUGGUUCCUCCUGGGACUAACGUUAAUCGCCGUUUCCAAUAAUGGUGAGUA----- ((((((..(((((((((...((((((((....)))))))).))))...)))))..)..)))))........----- ( -22.40, z-score = -1.64, R) >droWil1.scaffold_181130 11053171 76 + 16660200 AUAAUGGGGCGGCUAACUAUAUGGUUCCUAUUGGGACUAACGUUAAUCGCCGUCUCCAACAAUGGUGAGUUUAAAA ....(((((((((((((....(((((((....)))))))..))))...)))))).))).................. ( -23.40, z-score = -2.62, R) >droVir3.scaffold_12822 1579932 70 + 4096053 AUUAUGGGGCGGCUAACAAUUUGGUUCCUGCUGGGACUUACGUUAAUCGCCGUUUCCAACAAUGGUGAGU------ .......((((..((((.....((((((....))))))...))))..))))....(((....))).....------ ( -20.30, z-score = -1.23, R) >droMoj3.scaffold_6540 22614813 71 - 34148556 AUUAUGGGGCGUUUAACACUUUGGUUCCUGCUGGGACUGACGUUAAUCGCCGUUUCCUAUAAUGGUGAGUG----- (((((((((((.(((((...(..(((((....)))))..).))))).))).....))))))))........----- ( -21.90, z-score = -2.25, R) >droGri2.scaffold_14624 2621811 71 + 4233967 AUUAUGGGGCGACUAACAAUUUGGUUCCUGCUGGGACUGACGUUAAUCGCCGUAUCCAAUAAUGGUGAGUG----- ...(((.(((((.((((...(..(((((....)))))..).)))).))))).)))(((....)))......----- ( -22.70, z-score = -2.37, R) >anoGam1.chr2R 41154085 71 - 62725911 ACCAUGCGACGACUUUCAGUGUGGGUCCUGUUGGGCCUAGCGCUGUUUACCCUAAGCACAGCGGGUAAGUA----- ....(((.........((((((((((((....))))))).))))).((((((..........)))))))))----- ( -24.30, z-score = -0.70, R) >consensus AUUAUGGGGCGGCUAACAAUUUGGUUCCUGCUGGGACUAACGUUAAUCGCCGUUUCCAAUAAUGGUGAGUA_____ ....(((((((..((((...((((((((....)))))))).))))..))))....))).................. (-19.51 = -19.46 + -0.04)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:13:08 2011