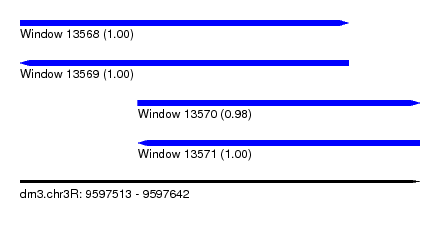
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,597,513 – 9,597,642 |
| Length | 129 |
| Max. P | 0.999599 |

| Location | 9,597,513 – 9,597,619 |
|---|---|
| Length | 106 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 84.78 |
| Shannon entropy | 0.31544 |
| G+C content | 0.43348 |
| Mean single sequence MFE | -34.62 |
| Consensus MFE | -27.46 |
| Energy contribution | -27.03 |
| Covariance contribution | -0.43 |
| Combinations/Pair | 1.16 |
| Mean z-score | -5.06 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.06 |
| SVM RNA-class probability | 0.999599 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
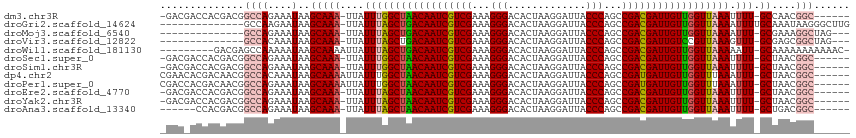
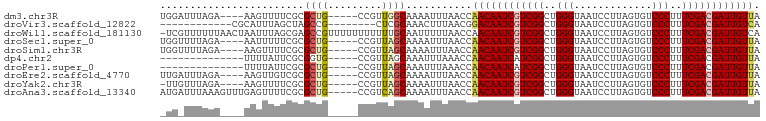
>dm3.chr3R 9597513 106 + 27905053 -GACGACCACGACGGCCAGAAAUAAGCAAA-UUAUUUGGCUAACAAUCGUCGAAAGGGACACUAAGGAUUACCCAGCCGACGAUUGUUGGUUAAAUUUU-GCCAACGGC------ -.............((((....)..(((((-..(((((((((((((((((((...(((.............)))...))))))))))))))))))))))-))....)))------ ( -34.52, z-score = -4.28, R) >droGri2.scaffold_14624 2621181 100 + 4233967 --------------GCCAAGAAUAAGCAAA-UUAUUUAGCUGACAAUCGUCGAAAGGGACACUAAGGAUUACCCAGCCGACGAUUGUUGGUUAAAAUUUUGCAAAUAAGGGCUUG --------------(((........(((((-...((((((..((((((((((...(((.............)))...))))))))))..))))))..))))).......)))... ( -36.58, z-score = -5.98, R) >droMoj3.scaffold_6540 22614213 96 - 34148556 --------------GCCAGAAAUAAGCAAA-UUAUUUAGCUGACAAUCGUCGAAAGGGACACUAAGGAUUACCCAGCCGACGAUUGUUGGUUAAAAUUU-GCGAAAGGCUAG--- --------------((((....)..(((((-((..(((((..((((((((((...(((.............)))...))))))))))..))))))))))-))....)))...--- ( -36.42, z-score = -6.38, R) >droVir3.scaffold_12822 1579435 96 + 4096053 --------------GCCACAAAUAAGCAAA-UUAUUUAGCUGACAAUCGUCGAAAGGGACACUAAGGAUUACCCAGCCGACGAUUGUCCGUUAAAGUUU-GCGAGCGGCUAG--- --------------(((.(......(((((-(..((((((.(((((((((((...(((.............)))...))))))))))).))))))))))-))..).)))...--- ( -35.22, z-score = -5.28, R) >droWil1.scaffold_181130 11052540 104 + 16660200 ---------GACGAGCCAAAAAUAAGCAAAAUUAUUUAGCUGACAAUCGUCGAAAGGGACACUAAGGAUUACCCAGCCGACGAUUGUUGGUUAAAAAUU-GCAAAAAAAAAAAC- ---------................((((.....((((((..((((((((((...(((.............)))...))))))))))..))))))..))-))............- ( -29.92, z-score = -5.46, R) >droSec1.super_0 8710751 106 - 21120651 -GACGACCACGACGGCCAGAAAUAAGCAAA-UUAUUUGGCUAACAAUCGUCGAAAGGGACACUAAGGAUUACCCAGCCGACGAUUGUUGGUUAAAUUUU-GCUAACGGC------ -.............((((....).((((((-..(((((((((((((((((((...(((.............)))...))))))))))))))))))))))-)))...)))------ ( -36.12, z-score = -4.97, R) >droSim1.chr3R 15688400 106 - 27517382 -GACGACCACGACGGCCAGAAAUAAGCAAA-UUAUUUGGCUAACAAUCGUCGAAAGGGACACUAAGGAUUACCCAGCCGACGAUUGUUGGUUAAAUUUU-GCUAACGGC------ -.............((((....).((((((-..(((((((((((((((((((...(((.............)))...))))))))))))))))))))))-)))...)))------ ( -36.12, z-score = -4.97, R) >dp4.chr2 10146010 108 + 30794189 CGAACACGACAACGGCCACAAAUAAGCAAAAUUAUUUGGCUAACAAUCGUCGAAAGGGACACUAAGGAUUACCCAGCCGAUGAUUGUUGGUUUAAAUUU-GCUAACGGC------ ..............(((.(((((((......)))))))((((((((((((((...(((.............)))...))))))))))))))........-......)))------ ( -30.52, z-score = -3.50, R) >droPer1.super_0 1487687 108 - 11822988 CGACCACGACAACGGCCAGAAAUAAGCAAAAUUAUUUGGCUAACAAUCGUCGAAAGGGACACUAAGGAUUACCCAGCCGAUGAUUGUUGGUUUAAAUUU-GCUAACGGC------ ..............((((....).((((((.(((...(((((((((((((((...(((.............)))...)))))))))))))))))).)))-)))...)))------ ( -30.52, z-score = -3.11, R) >droEre2.scaffold_4770 5711164 106 - 17746568 -GACGACCACGACGGCCAGAAAUAAGCAAA-UUAUUUAGCUAACAAUCGUCGAAAGGGACACUAAGGAUUACCCAGCCGACGAUUGUUGGUUAAAUUUU-GCUAACGGC------ -.............((((....).((((((-..(((((((((((((((((((...(((.............)))...))))))))))))))))))))))-)))...)))------ ( -36.52, z-score = -5.60, R) >droYak2.chr3R 13906049 106 + 28832112 -GACGACCACGACGGCCAGAAAUAAGCAAA-UUAUUUAGCUAACAAUCGUCGAAAGGGACACUAAGGAUUACCCAGCCGACGAUUGUUGGUUAAAUUUU-GCUAACGGC------ -.............((((....).((((((-..(((((((((((((((((((...(((.............)))...))))))))))))))))))))))-)))...)))------ ( -36.52, z-score = -5.60, R) >droAna3.scaffold_13340 17923125 101 + 23697760 ------CCACGACGGCCAGAAAUAAGCAAA-UUAUUUAGCUAACAAUCGUCGAAAGGGACACUAAGGAUUACCCAGCCGACGAUUGUUGGUUAAAUUUU-GCUGACGGC------ ------........(((.......((((((-..(((((((((((((((((((...(((.............)))...))))))))))))))))))))))-)))...)))------ ( -36.52, z-score = -5.59, R) >consensus _GAC_ACCACGACGGCCAGAAAUAAGCAAA_UUAUUUAGCUAACAAUCGUCGAAAGGGACACUAAGGAUUACCCAGCCGACGAUUGUUGGUUAAAUUUU_GCUAACGGC______ .........................((.((....((((((((((((((((((...(((.............)))...))))))))))))))))))..)).))............. (-27.46 = -27.03 + -0.43)
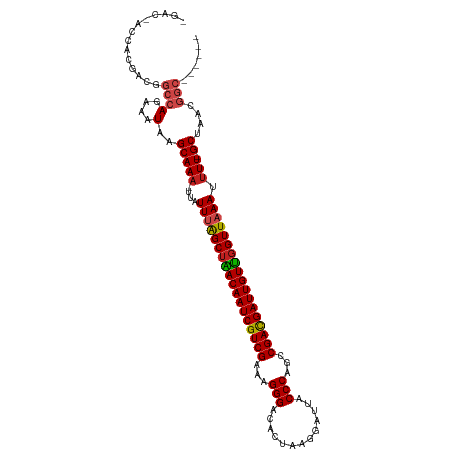
| Location | 9,597,513 – 9,597,619 |
|---|---|
| Length | 106 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 84.78 |
| Shannon entropy | 0.31544 |
| G+C content | 0.43348 |
| Mean single sequence MFE | -29.84 |
| Consensus MFE | -23.71 |
| Energy contribution | -23.77 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.29 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.27 |
| SVM RNA-class probability | 0.998162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
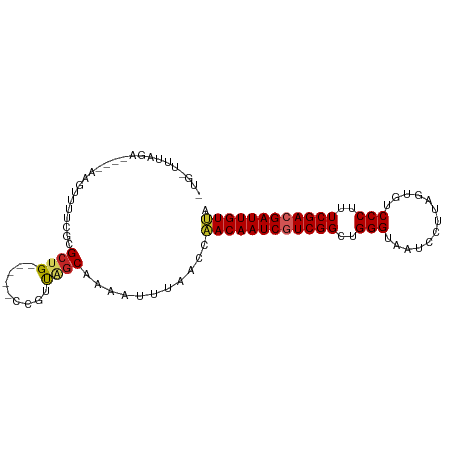
>dm3.chr3R 9597513 106 - 27905053 ------GCCGUUGGC-AAAAUUUAACCAACAAUCGUCGGCUGGGUAAUCCUUAGUGUCCCUUUCGACGAUUGUUAGCCAAAUAA-UUUGCUUAUUUCUGGCCGUCGUGGUCGUC- ------(((...)))-........(((((((((((((((..(((.............)))..)))))))))))).(((((((((-.....)))))..))))......)))....- ( -31.52, z-score = -2.81, R) >droGri2.scaffold_14624 2621181 100 - 4233967 CAAGCCCUUAUUUGCAAAAUUUUAACCAACAAUCGUCGGCUGGGUAAUCCUUAGUGUCCCUUUCGACGAUUGUCAGCUAAAUAA-UUUGCUUAUUCUUGGC-------------- ...(((...((..(((((..((((.(..(((((((((((..(((.............)))..)))))))))))..).))))...-)))))..))....)))-------------- ( -29.82, z-score = -4.50, R) >droMoj3.scaffold_6540 22614213 96 + 34148556 ---CUAGCCUUUCGC-AAAUUUUAACCAACAAUCGUCGGCUGGGUAAUCCUUAGUGUCCCUUUCGACGAUUGUCAGCUAAAUAA-UUUGCUUAUUUCUGGC-------------- ---((((......((-((((((((.(..(((((((((((..(((.............)))..)))))))))))..).)))..))-)))))......)))).-------------- ( -28.02, z-score = -4.12, R) >droVir3.scaffold_12822 1579435 96 - 4096053 ---CUAGCCGCUCGC-AAACUUUAACGGACAAUCGUCGGCUGGGUAAUCCUUAGUGUCCCUUUCGACGAUUGUCAGCUAAAUAA-UUUGCUUAUUUGUGGC-------------- ---...(((((..((-(((.((((.(.((((((((((((..(((.............)))..)))))))))))).).))))...-)))))......)))))-------------- ( -34.42, z-score = -4.50, R) >droWil1.scaffold_181130 11052540 104 - 16660200 -GUUUUUUUUUUUGC-AAUUUUUAACCAACAAUCGUCGGCUGGGUAAUCCUUAGUGUCCCUUUCGACGAUUGUCAGCUAAAUAAUUUUGCUUAUUUUUGGCUCGUC--------- -(((.........((-((..((((.(..(((((((((((..(((.............)))..)))))))))))..).)))).....))))........))).....--------- ( -24.55, z-score = -3.13, R) >droSec1.super_0 8710751 106 + 21120651 ------GCCGUUAGC-AAAAUUUAACCAACAAUCGUCGGCUGGGUAAUCCUUAGUGUCCCUUUCGACGAUUGUUAGCCAAAUAA-UUUGCUUAUUUCUGGCCGUCGUGGUCGUC- ------......(((-(((((((..(.((((((((((((..(((.............)))..)))))))))))).)..))))..-)))))).......((((.....))))...- ( -30.72, z-score = -3.03, R) >droSim1.chr3R 15688400 106 + 27517382 ------GCCGUUAGC-AAAAUUUAACCAACAAUCGUCGGCUGGGUAAUCCUUAGUGUCCCUUUCGACGAUUGUUAGCCAAAUAA-UUUGCUUAUUUCUGGCCGUCGUGGUCGUC- ------......(((-(((((((..(.((((((((((((..(((.............)))..)))))))))))).)..))))..-)))))).......((((.....))))...- ( -30.72, z-score = -3.03, R) >dp4.chr2 10146010 108 - 30794189 ------GCCGUUAGC-AAAUUUAAACCAACAAUCAUCGGCUGGGUAAUCCUUAGUGUCCCUUUCGACGAUUGUUAGCCAAAUAAUUUUGCUUAUUUGUGGCCGUUGUCGUGUUCG ------((((.((((-...........(((((((.((((..(((.............)))..)))).))))))).(((((((((......)))))..)))).)))).)).))... ( -25.22, z-score = -1.42, R) >droPer1.super_0 1487687 108 + 11822988 ------GCCGUUAGC-AAAUUUAAACCAACAAUCAUCGGCUGGGUAAUCCUUAGUGUCCCUUUCGACGAUUGUUAGCCAAAUAAUUUUGCUUAUUUCUGGCCGUUGUCGUGGUCG ------((((.((((-...........(((((((.((((..(((.............)))..)))).))))))).(((((((((......)))))..)))).))))...)))).. ( -25.72, z-score = -1.38, R) >droEre2.scaffold_4770 5711164 106 + 17746568 ------GCCGUUAGC-AAAAUUUAACCAACAAUCGUCGGCUGGGUAAUCCUUAGUGUCCCUUUCGACGAUUGUUAGCUAAAUAA-UUUGCUUAUUUCUGGCCGUCGUGGUCGUC- ------......(((-((((((((.(.((((((((((((..(((.............)))..)))))))))))).).)))))..-)))))).......((((.....))))...- ( -32.82, z-score = -3.83, R) >droYak2.chr3R 13906049 106 - 28832112 ------GCCGUUAGC-AAAAUUUAACCAACAAUCGUCGGCUGGGUAAUCCUUAGUGUCCCUUUCGACGAUUGUUAGCUAAAUAA-UUUGCUUAUUUCUGGCCGUCGUGGUCGUC- ------......(((-((((((((.(.((((((((((((..(((.............)))..)))))))))))).).)))))..-)))))).......((((.....))))...- ( -32.82, z-score = -3.83, R) >droAna3.scaffold_13340 17923125 101 - 23697760 ------GCCGUCAGC-AAAAUUUAACCAACAAUCGUCGGCUGGGUAAUCCUUAGUGUCCCUUUCGACGAUUGUUAGCUAAAUAA-UUUGCUUAUUUCUGGCCGUCGUGG------ ------((((..(((-((((((((.(.((((((((((((..(((.............)))..)))))))))))).).)))))..-))))))......))))........------ ( -31.72, z-score = -3.89, R) >consensus ______GCCGUUAGC_AAAAUUUAACCAACAAUCGUCGGCUGGGUAAUCCUUAGUGUCCCUUUCGACGAUUGUUAGCUAAAUAA_UUUGCUUAUUUCUGGCCGUCGUGGU_GUC_ ...........................((((((((((((..(((.............)))..)))))))))))).(((((((((......)))))..)))).............. (-23.71 = -23.77 + 0.06)


| Location | 9,597,551 – 9,597,642 |
|---|---|
| Length | 91 |
| Sequences | 10 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 81.48 |
| Shannon entropy | 0.38215 |
| G+C content | 0.42708 |
| Mean single sequence MFE | -22.67 |
| Consensus MFE | -19.34 |
| Energy contribution | -19.55 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.984005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9597551 91 + 27905053 UAACAAUCGUCGAAAGGGACACUAAGGAUUACCCAGCCGACGAUUGUUGGUUAAAUUUUGCCAACGG-----CAGCGCGAAAACUU----UCUAAAUCCA ((((((((((((...(((.............)))...))))))))))))........(((((...))-----)))...........----.......... ( -23.12, z-score = -1.71, R) >droVir3.scaffold_12822 1579460 80 + 4096053 UGACAAUCGUCGAAAGGGACACUAAGGAUUACCCAGCCGACGAUUGUCCGUUAAAGUUUGCGAG--------CGGCUAGCUAAAUGCG------------ .(((((((((((...(((.............)))...)))))))))))(((...((((.((...--------..)).))))....)))------------ ( -25.52, z-score = -2.28, R) >droWil1.scaffold_181130 11052571 99 + 16660200 UGACAAUCGUCGAAAGGGACACUAAGGAUUACCCAGCCGACGAUUGUUGGUUAAAAAUUGCAAAAAAAAAAACGGCUCGCUAAAUUAGUUAAAAAACGA- .(((((((((((...(((.............)))...)))))))))))...................................................- ( -18.72, z-score = -0.78, R) >droSec1.super_0 8710789 91 - 21120651 UAACAAUCGUCGAAAGGGACACUAAGGAUUACCCAGCCGACGAUUGUUGGUUAAAUUUUGCUAACGG-----CAGCGCGAAAAAUU----UCUAAAACCA .(((((((((((...(((.............)))...)))))))))))((((.....((((.....)-----)))...(((....)----))...)))). ( -24.32, z-score = -2.45, R) >droSim1.chr3R 15688438 91 - 27517382 UAACAAUCGUCGAAAGGGACACUAAGGAUUACCCAGCCGACGAUUGUUGGUUAAAUUUUGCUAACGG-----CAGCGCGAAAACUU----UCUAAAACCA .(((((((((((...(((.............)))...)))))))))))((((.....((((.....)-----)))...(((....)----))...)))). ( -24.32, z-score = -2.37, R) >dp4.chr2 10146050 81 + 30794189 UAACAAUCGUCGAAAGGGACACUAAGGAUUACCCAGCCGAUGAUUGUUGGUUUAAAUUUGCUAACGG-----CACCGCGAAUAAAA-------------- ((((((((((((...(((.............)))...))))))))))))......((((((......-----....))))))....-------------- ( -20.92, z-score = -2.05, R) >droPer1.super_0 1487727 81 - 11822988 UAACAAUCGUCGAAAGGGACACUAAGGAUUACCCAGCCGAUGAUUGUUGGUUUAAAUUUGCUAACGG-----CAGCGCGAAUAAAA-------------- ((((((((((((...(((.............)))...))))))))))))......((((((......-----....))))))....-------------- ( -20.92, z-score = -1.95, R) >droEre2.scaffold_4770 5711202 91 - 17746568 UAACAAUCGUCGAAAGGGACACUAAGGAUUACCCAGCCGACGAUUGUUGGUUAAAUUUUGCUAACGG-----CAGCGCGACAACUU----UCUAAAUCAA ((((((((((((...(((.............)))...))))))))))))(((.....((((.....)-----)))...))).....----.......... ( -22.02, z-score = -1.67, R) >droYak2.chr3R 13906087 90 + 28832112 UAACAAUCGUCGAAAGGGACACUAAGGAUUACCCAGCCGACGAUUGUUGGUUAAAUUUUGCUAACGG-----CAGCGCGAAAACUU----UCUAAACAA- ((((((((((((...(((.............)))...))))))))))))......((((((......-----....))))))....----.........- ( -22.62, z-score = -1.94, R) >droAna3.scaffold_13340 17923158 95 + 23697760 UAACAAUCGUCGAAAGGGACACUAAGGAUUACCCAGCCGACGAUUGUUGGUUAAAUUUUGCUGACGG-----CAGCGCGAAAACUCAAACUUUAAAUCAU ((((((((((((...(((.............)))...))))))))))))..........((((....-----))))........................ ( -24.22, z-score = -2.22, R) >consensus UAACAAUCGUCGAAAGGGACACUAAGGAUUACCCAGCCGACGAUUGUUGGUUAAAUUUUGCUAACGG_____CAGCGCGAAAAAUU____UCUAAA_CA_ ((((((((((((...(((.............)))...))))))))))))......((((((...............)))))).................. (-19.34 = -19.55 + 0.21)
| Location | 9,597,551 – 9,597,642 |
|---|---|
| Length | 91 |
| Sequences | 10 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 81.48 |
| Shannon entropy | 0.38215 |
| G+C content | 0.42708 |
| Mean single sequence MFE | -24.65 |
| Consensus MFE | -21.30 |
| Energy contribution | -21.58 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.77 |
| SVM RNA-class probability | 0.995147 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9597551 91 - 27905053 UGGAUUUAGA----AAGUUUUCGCGCUG-----CCGUUGGCAAAAUUUAACCAACAAUCGUCGGCUGGGUAAUCCUUAGUGUCCCUUUCGACGAUUGUUA .((.....((----(....)))....((-----((...))))........))((((((((((((..(((.............)))..)))))))))))). ( -26.82, z-score = -2.06, R) >droVir3.scaffold_12822 1579460 80 - 4096053 ------------CGCAUUUAGCUAGCCG--------CUCGCAAACUUUAACGGACAAUCGUCGGCUGGGUAAUCCUUAGUGUCCCUUUCGACGAUUGUCA ------------.((....(((.....)--------)).))...........((((((((((((..(((.............)))..)))))))))))). ( -26.82, z-score = -3.03, R) >droWil1.scaffold_181130 11052571 99 - 16660200 -UCGUUUUUUAACUAAUUUAGCGAGCCGUUUUUUUUUUUGCAAUUUUUAACCAACAAUCGUCGGCUGGGUAAUCCUUAGUGUCCCUUUCGACGAUUGUCA -(((((.............))))).............................(((((((((((..(((.............)))..))))))))))).. ( -22.14, z-score = -2.23, R) >droSec1.super_0 8710789 91 + 21120651 UGGUUUUAGA----AAUUUUUCGCGCUG-----CCGUUAGCAAAAUUUAACCAACAAUCGUCGGCUGGGUAAUCCUUAGUGUCCCUUUCGACGAUUGUUA .((((...((----(....)))..((((-----....)))).......))))((((((((((((..(((.............)))..)))))))))))). ( -27.72, z-score = -3.22, R) >droSim1.chr3R 15688438 91 + 27517382 UGGUUUUAGA----AAGUUUUCGCGCUG-----CCGUUAGCAAAAUUUAACCAACAAUCGUCGGCUGGGUAAUCCUUAGUGUCCCUUUCGACGAUUGUUA .((((...((----(....)))..((((-----....)))).......))))((((((((((((..(((.............)))..)))))))))))). ( -27.72, z-score = -2.96, R) >dp4.chr2 10146050 81 - 30794189 --------------UUUUAUUCGCGGUG-----CCGUUAGCAAAUUUAAACCAACAAUCAUCGGCUGGGUAAUCCUUAGUGUCCCUUUCGACGAUUGUUA --------------........(((...-----.)))...............(((((((.((((..(((.............)))..)))).))))))). ( -17.82, z-score = -0.97, R) >droPer1.super_0 1487727 81 + 11822988 --------------UUUUAUUCGCGCUG-----CCGUUAGCAAAUUUAAACCAACAAUCAUCGGCUGGGUAAUCCUUAGUGUCCCUUUCGACGAUUGUUA --------------..........((((-----....))))...........(((((((.((((..(((.............)))..)))).))))))). ( -18.82, z-score = -1.77, R) >droEre2.scaffold_4770 5711202 91 + 17746568 UUGAUUUAGA----AAGUUGUCGCGCUG-----CCGUUAGCAAAAUUUAACCAACAAUCGUCGGCUGGGUAAUCCUUAGUGUCCCUUUCGACGAUUGUUA .....(((((----...((((.(((...-----.)))..))))..)))))..((((((((((((..(((.............)))..)))))))))))). ( -24.62, z-score = -2.07, R) >droYak2.chr3R 13906087 90 - 28832112 -UUGUUUAGA----AAGUUUUCGCGCUG-----CCGUUAGCAAAAUUUAACCAACAAUCGUCGGCUGGGUAAUCCUUAGUGUCCCUUUCGACGAUUGUUA -(((((..(.----.(((......))).-----.)...))))).........((((((((((((..(((.............)))..)))))))))))). ( -25.12, z-score = -2.52, R) >droAna3.scaffold_13340 17923158 95 - 23697760 AUGAUUUAAAGUUUGAGUUUUCGCGCUG-----CCGUCAGCAAAAUUUAACCAACAAUCGUCGGCUGGGUAAUCCUUAGUGUCCCUUUCGACGAUUGUUA ..........(.(((((((((...((((-----....))))))))))))).)((((((((((((..(((.............)))..)))))))))))). ( -28.92, z-score = -3.41, R) >consensus _UG_UUUAGA____AAGUUUUCGCGCUG_____CCGUUAGCAAAAUUUAACCAACAAUCGUCGGCUGGGUAAUCCUUAGUGUCCCUUUCGACGAUUGUUA ........................((((.........))))...........((((((((((((..(((.............)))..)))))))))))). (-21.30 = -21.58 + 0.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:13:06 2011