| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,597,285 – 9,597,380 |
| Length | 95 |
| Max. P | 0.897952 |

| Location | 9,597,285 – 9,597,380 |
|---|---|
| Length | 95 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.77 |
| Shannon entropy | 0.27221 |
| G+C content | 0.43411 |
| Mean single sequence MFE | -25.93 |
| Consensus MFE | -25.30 |
| Energy contribution | -25.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.57 |
| Structure conservation index | 0.98 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.618908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
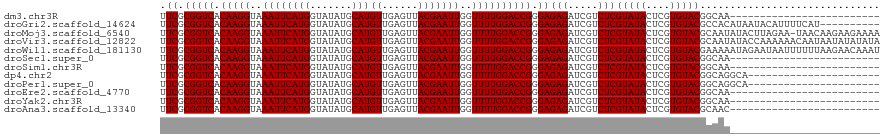
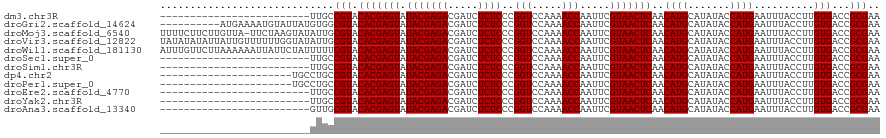
>dm3.chr3R 9597285 95 + 27905053 UUCGCGGUCACAAGGUAAAUUCAUGGUAUAUGCAUGUUGAGUUACGAAUUGGUUUUGGACCGGGAGAGAUCGUCUCGUAUACUCGUGUACGGCAA------------------------- .((.(((((.(((((..((((((((.......)))((......)))))))..)))))))))).))(((.....)))(((((....))))).....------------------------- ( -25.30, z-score = -0.62, R) >droGri2.scaffold_14624 2620964 110 + 4233967 UUCGCGGUCACAAGGUAAAUUCAUGGUAUAUGCAUGUUGAGUUACGAAUUGGUUUUGGACCGGGAGAGAUCGUCUCGUAUACUCGUGUACGCCACAUAAUACAUUUUCAU---------- (((.(((((.(((((..((((((((.......)))((......)))))))..)))))))))).))).....((..((((((....))))))..))...............---------- ( -25.50, z-score = -0.01, R) >droMoj3.scaffold_6540 22613972 119 - 34148556 UUCGCGGUCACAAGGUAAAUUCAUGGUAUAUGCAUGUUGAGUUACGAAUUGGUUUUGGACCGGGAGAGAUCGUCUCGUAUACUCGUGUACGCAAUAUACUUAGAA-UAACAAGAAGAAAA .((.(((((.(((((..((((((((.......)))((......)))))))..)))))))))).))(((.....)))(((((....)))))...............-.............. ( -25.30, z-score = 0.08, R) >droVir3.scaffold_12822 1579195 120 + 4096053 UUCGCGGUCACAAGGUAAAUUCAUGGUAUAUGCAUGUUGAGUUACGAAUUGGUUUUGGACCGGGAGAGAUCGUCUCGUAUACUCGUGUACGCAAUAUACCAAAAAACAAUAAUAUAUAUA .......................((((((((((....((((((((((..(((((((.........)))))))..))))).))))).....)).))))))))................... ( -25.70, z-score = -0.08, R) >droWil1.scaffold_181130 11052269 120 + 16660200 UUCGCGGUCACAAGGUAAAUUCAUGGUAUAUGCAUGUUGAGUUACGAAUUGGUUUUGGACCGGGAGAGAUCGUCUCGUAUACUCGUGUACGAAAAAUAGAAUAAUUUUUUAAGAACAAAU .((.(((((.(((((..((((((((.......)))((......)))))))..)))))))))).))(((.....)))(((((....)))))(((((((......))))))).......... ( -26.90, z-score = -0.83, R) >droSec1.super_0 8710523 95 - 21120651 UUCGCGGUCACAAGGUAAAUUCAUGGUAUAUGCAUGUUGAGUUACGAAUUGGUUUUGGACCGGGAGAGAUCGUCUCGUAUACUCGUGUACGGCAA------------------------- .((.(((((.(((((..((((((((.......)))((......)))))))..)))))))))).))(((.....)))(((((....))))).....------------------------- ( -25.30, z-score = -0.62, R) >droSim1.chr3R 15688172 95 - 27517382 UUCGCGGUCACAAGGUAAAUUCAUGGUAUAUGCAUGUUGAGUUACGAAUUGGUUUUGGACCGGGAGAGAUCGUCUCGUAUACUCGUGUACGGCAA------------------------- .((.(((((.(((((..((((((((.......)))((......)))))))..)))))))))).))(((.....)))(((((....))))).....------------------------- ( -25.30, z-score = -0.62, R) >dp4.chr2 10145769 98 + 30794189 UUCGCGGUCACAAGGUAAAUUCAUGGUAUAUGCAUGUUGAGUUACGAAUUGGUUUUGGACCGGGAGAGAUCGUCUCGUAUACUCGUGUACGGCAGGCA---------------------- (((.(((((.(((((..((((((((.......)))((......)))))))..)))))))))).))).....((((((((((....))))))..)))).---------------------- ( -28.00, z-score = -1.10, R) >droPer1.super_0 1487445 98 - 11822988 UUCGCGGUCACAAGGUAAAUUCAUGGUAUAUGCAUGUUGAGUUACGAAUUGGUUUUGGACCGGGAGAGAUCGUCUCGUAUACUCGUGUACGGCAGGCA---------------------- (((.(((((.(((((..((((((((.......)))((......)))))))..)))))))))).))).....((((((((((....))))))..)))).---------------------- ( -28.00, z-score = -1.10, R) >droEre2.scaffold_4770 5710937 95 - 17746568 UUCGCGGUCACAAGGUAAAUUCAUGGUAUAUGCAUGUUGAGUUACGAAUUGGUUUUGGACCGGGAGAGAUCGUCUCGUAUACUCGUGUACGGCAA------------------------- .((.(((((.(((((..((((((((.......)))((......)))))))..)))))))))).))(((.....)))(((((....))))).....------------------------- ( -25.30, z-score = -0.62, R) >droYak2.chr3R 13905826 95 + 28832112 UUCGCGGUCACAAGGUAAAUUCAUGGUAUAUGCAUGUUGAGUUACGAAUUGGUUUUGGACCGGGAGAGAUCGUCUCGUAUACUCGUGUACGGCAA------------------------- .((.(((((.(((((..((((((((.......)))((......)))))))..)))))))))).))(((.....)))(((((....))))).....------------------------- ( -25.30, z-score = -0.62, R) >droAna3.scaffold_13340 17922906 95 + 23697760 UUCGCGGUCACAAGGUAAAUUCAUGGUAUAUGCAUGUUGAGUUACGAAUUGGUUUUGGACCGGGAGAGAUCGUCUCGUAUACUCGUGUACGCAAC------------------------- .((.(((((.(((((..((((((((.......)))((......)))))))..)))))))))).))(((.....)))(((((....))))).....------------------------- ( -25.30, z-score = -0.72, R) >consensus UUCGCGGUCACAAGGUAAAUUCAUGGUAUAUGCAUGUUGAGUUACGAAUUGGUUUUGGACCGGGAGAGAUCGUCUCGUAUACUCGUGUACGGCAA_________________________ .((.(((((.(((((..((((((((.......)))((......)))))))..)))))))))).))(((.....)))(((((....))))).............................. (-25.30 = -25.30 + 0.00)
| Location | 9,597,285 – 9,597,380 |
|---|---|
| Length | 95 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.77 |
| Shannon entropy | 0.27221 |
| G+C content | 0.43411 |
| Mean single sequence MFE | -21.55 |
| Consensus MFE | -17.35 |
| Energy contribution | -17.35 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.897952 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
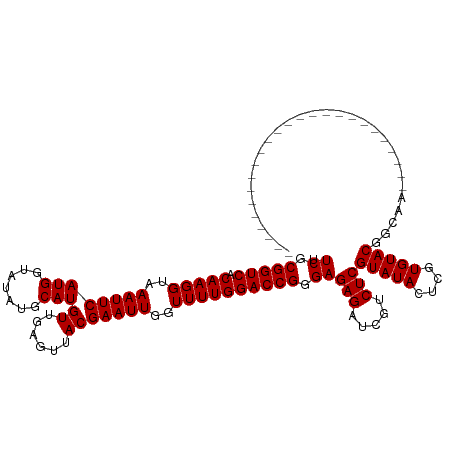
>dm3.chr3R 9597285 95 - 27905053 -------------------------UUGCCGUACACGAGUAUACGAGACGAUCUCUCCCGGUCCAAAACCAAUUCGUAACUCAACAUGCAUAUACCAUGAAUUUACCUUGUGACCGCGAA -------------------------((((.((.(((((((.(((((((.....))))..(((.....))).....)))))))..((((.......))))..........))))).)))). ( -21.30, z-score = -2.32, R) >droGri2.scaffold_14624 2620964 110 - 4233967 ----------AUGAAAAUGUAUUAUGUGGCGUACACGAGUAUACGAGACGAUCUCUCCCGGUCCAAAACCAAUUCGUAACUCAACAUGCAUAUACCAUGAAUUUACCUUGUGACCGCGAA ----------..................(((..(((((((.(((((((.....))))..(((.....))).....)))))))..((((.......))))..........)))..)))... ( -21.20, z-score = -0.25, R) >droMoj3.scaffold_6540 22613972 119 + 34148556 UUUUCUUCUUGUUA-UUCUAAGUAUAUUGCGUACACGAGUAUACGAGACGAUCUCUCCCGGUCCAAAACCAAUUCGUAACUCAACAUGCAUAUACCAUGAAUUUACCUUGUGACCGCGAA ...((.((((((((-(((...((((.....))))..))))).)))))).))....((.((((((((....(((((((...................)))))))....))).))))).)). ( -23.71, z-score = -1.81, R) >droVir3.scaffold_12822 1579195 120 - 4096053 UAUAUAUAUUAUUGUUUUUUGGUAUAUUGCGUACACGAGUAUACGAGACGAUCUCUCCCGGUCCAAAACCAAUUCGUAACUCAACAUGCAUAUACCAUGAAUUUACCUUGUGACCGCGAA ....(((((((........)))))))(((((..(((((((.(((((((.....))))..(((.....))).....)))))))..((((.......))))..........)))..))))). ( -25.30, z-score = -1.52, R) >droWil1.scaffold_181130 11052269 120 - 16660200 AUUUGUUCUUAAAAAAUUAUUCUAUUUUUCGUACACGAGUAUACGAGACGAUCUCUCCCGGUCCAAAACCAAUUCGUAACUCAACAUGCAUAUACCAUGAAUUUACCUUGUGACCGCGAA ...........................(((((.(((((((.(((((((.....))))..(((.....))).....)))))))..((((.......))))..........)))...))))) ( -19.00, z-score = -0.91, R) >droSec1.super_0 8710523 95 + 21120651 -------------------------UUGCCGUACACGAGUAUACGAGACGAUCUCUCCCGGUCCAAAACCAAUUCGUAACUCAACAUGCAUAUACCAUGAAUUUACCUUGUGACCGCGAA -------------------------((((.((.(((((((.(((((((.....))))..(((.....))).....)))))))..((((.......))))..........))))).)))). ( -21.30, z-score = -2.32, R) >droSim1.chr3R 15688172 95 + 27517382 -------------------------UUGCCGUACACGAGUAUACGAGACGAUCUCUCCCGGUCCAAAACCAAUUCGUAACUCAACAUGCAUAUACCAUGAAUUUACCUUGUGACCGCGAA -------------------------((((.((.(((((((.(((((((.....))))..(((.....))).....)))))))..((((.......))))..........))))).)))). ( -21.30, z-score = -2.32, R) >dp4.chr2 10145769 98 - 30794189 ----------------------UGCCUGCCGUACACGAGUAUACGAGACGAUCUCUCCCGGUCCAAAACCAAUUCGUAACUCAACAUGCAUAUACCAUGAAUUUACCUUGUGACCGCGAA ----------------------....(((.((.(((((((.(((((((.....))))..(((.....))).....)))))))..((((.......))))..........))))).))).. ( -20.10, z-score = -1.67, R) >droPer1.super_0 1487445 98 + 11822988 ----------------------UGCCUGCCGUACACGAGUAUACGAGACGAUCUCUCCCGGUCCAAAACCAAUUCGUAACUCAACAUGCAUAUACCAUGAAUUUACCUUGUGACCGCGAA ----------------------....(((.((.(((((((.(((((((.....))))..(((.....))).....)))))))..((((.......))))..........))))).))).. ( -20.10, z-score = -1.67, R) >droEre2.scaffold_4770 5710937 95 + 17746568 -------------------------UUGCCGUACACGAGUAUACGAGACGAUCUCUCCCGGUCCAAAACCAAUUCGUAACUCAACAUGCAUAUACCAUGAAUUUACCUUGUGACCGCGAA -------------------------((((.((.(((((((.(((((((.....))))..(((.....))).....)))))))..((((.......))))..........))))).)))). ( -21.30, z-score = -2.32, R) >droYak2.chr3R 13905826 95 - 28832112 -------------------------UUGCCGUACACGAGUAUACGAGACGAUCUCUCCCGGUCCAAAACCAAUUCGUAACUCAACAUGCAUAUACCAUGAAUUUACCUUGUGACCGCGAA -------------------------((((.((.(((((((.(((((((.....))))..(((.....))).....)))))))..((((.......))))..........))))).)))). ( -21.30, z-score = -2.32, R) >droAna3.scaffold_13340 17922906 95 - 23697760 -------------------------GUUGCGUACACGAGUAUACGAGACGAUCUCUCCCGGUCCAAAACCAAUUCGUAACUCAACAUGCAUAUACCAUGAAUUUACCUUGUGACCGCGAA -------------------------.(((((..(((((((.(((((((.....))))..(((.....))).....)))))))..((((.......))))..........)))..))))). ( -22.70, z-score = -2.33, R) >consensus _________________________UUGCCGUACACGAGUAUACGAGACGAUCUCUCCCGGUCCAAAACCAAUUCGUAACUCAACAUGCAUAUACCAUGAAUUUACCUUGUGACCGCGAA .............................(((.(((((((.(((((((.....))))..(((.....))).....)))))))..((((.......))))..........)))...))).. (-17.35 = -17.35 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:13:03 2011