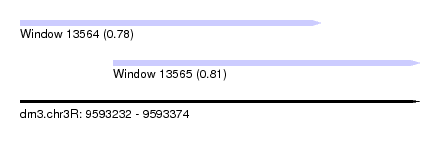
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,593,232 – 9,593,374 |
| Length | 142 |
| Max. P | 0.810972 |

| Location | 9,593,232 – 9,593,339 |
|---|---|
| Length | 107 |
| Sequences | 10 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 69.20 |
| Shannon entropy | 0.61727 |
| G+C content | 0.51680 |
| Mean single sequence MFE | -25.78 |
| Consensus MFE | -11.97 |
| Energy contribution | -11.39 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.775590 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
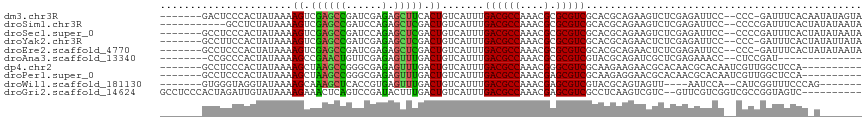
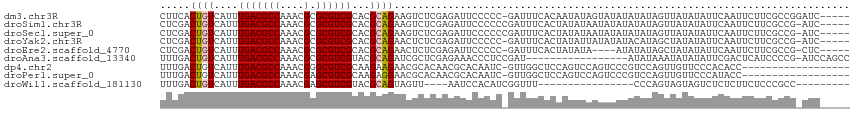
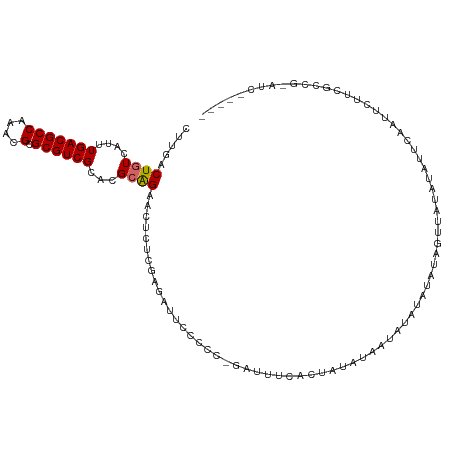
>dm3.chr3R 9593232 107 + 27905053 -------GACUCCCACUAUAAAAGUCGAGCCGAUCGAGAGCUUCACUGUCAUUUGACGCCAAACGCGCGUCGCACGCAGAAGUCUCGAGAUUCC--CCC-GAUUUCACAAUAUAGUA -------.......((((((.((((((.(..((((..(((((((..(((....(((((((....).))))))...))))))).)))..))))..--).)-))))).....)))))). ( -28.00, z-score = -2.22, R) >droSim1.chr3R 15684245 104 - 27517382 -----------GCCUCUAUAAAAGUCGAGCCGAUCCAGAGCUCGACUGUCAUUUGACGCCAAACGCGCGUCGCACGCAGAAGUCUCGAGAUUCC--CCCCGAUUUCACUAUAUAAUA -----------((.........(((((((((......).)))))))).......((((((....).))))))).....((((((..........--....))))))........... ( -27.04, z-score = -2.23, R) >droSec1.super_0 8705962 108 - 21120651 -------GCCUCCCACUAUAAAAGUCGAGCCGAUCCAGAGCUCGACUGUCAUUUGACGCCAAACGCGCGUCGCACGCAGAAGUCUCGAGAUUCC--CCCCGAUUUCACUAUAUAAUA -------((.............(((((((((......).)))))))).......((((((....).))))))).....((((((..........--....))))))........... ( -27.04, z-score = -2.53, R) >droYak2.chr3R 13901694 107 + 28832112 -------GCCUUCCACUAUAAAAGUCGAGCCGAUCGAGAGCUCGACUGUCAUUUGACGCCAAACGCGCGUCGCACGCAGAACUCUCGAGAUUCC--CCC-GAUUUCACUAUAUUAUA -------((.............(((((((((......).)))))))).......((((((....).))))))).............((((((..--...-))))))........... ( -26.10, z-score = -1.61, R) >droEre2.scaffold_4770 5706791 107 - 17746568 -------GCCUCCCACUAUAAAAGUCGAGCCGAUCGAGAGCUCGACUGUCAUUUGACGCCAAACGCGCGUCGCACGCAGAACUCUCGAGAUUCC--CCC-GAUUUCACUAUAUAAUA -------((.............(((((((((......).)))))))).......((((((....).))))))).............((((((..--...-))))))........... ( -26.10, z-score = -1.80, R) >droAna3.scaffold_13340 17919079 93 + 23697760 --------CCGCCCACUAUAAAAGCCGAACUGUUCGAGAGUUUGACUGUCAUUUGACGCCAAACGCGCGUCGUACGCAGAUCGCUCGAGAAACC--CUCCGAU-------------- --------.........................(((.((((..((((((....(((((((....).))))))...)))).))))))(((.....--)))))).-------------- ( -20.30, z-score = -0.14, R) >dp4.chr2 10142099 100 + 30794189 -------GCCUCCCACUAUAAAAGCUAAGCCGGGCGAGAGUUUGACUGUCAUUUGACGCCAAACGGGCGUCGCAAGAAGAACGCACAACGCACAAUCGUUGGCUCCA---------- -------................((..((.(((((....))))).)).......((((((.....)))))))).........((.(((((......)))))))....---------- ( -26.20, z-score = 0.16, R) >droPer1.super_0 1483834 100 - 11822988 -------GCCUCCCACUAUAAAAGCUAAGCCGGGCGAGAGUUUGACUGUCAUUUGACGCCAAACGAGCGUCGCAAGAGGAACGCACAACGCACAAUCGUUGGCUCCA---------- -------.((((........(((....((.(((((....))))).))....)))((((((....).)))))....))))...((.(((((......)))))))....---------- ( -26.10, z-score = 0.01, R) >droWil1.scaffold_181130 11047310 97 + 16660200 -------GUGGGUAGGUAUAAAAGCAAAGCUCACCGUGAGUUUGACUGUCAUUUGACGCCAAACGAGCGUCGUACGCAGUAGUU----AAUCCA--CAUCGGUUUCCCAG------- -------((((((.............((((((.....)))))).(((((....(((((((....).))))))...)))))....----.)))))--)...((....))..------- ( -27.30, z-score = -1.73, R) >droGri2.scaffold_14624 2615678 105 + 4233967 GCCUCCCACUAGAUUGUAUAAAAGAAACUCAGUCCGAUACUUUGACUGUCAUUUGACGCCAAACGAGCGUCGCCUCAAGUCGUC--GUUCGUCGGUCGCCGGUAGUC---------- (((....((......))..............(.(((((((..(((((......(((((((....).)))))).....)))))..--))..))))).)...)))....---------- ( -23.60, z-score = 0.13, R) >consensus _______GCCUCCCACUAUAAAAGUCGAGCCGAUCGAGAGCUUGACUGUCAUUUGACGCCAAACGCGCGUCGCACGCAGAACUCUCGAGAUUCC__CCCCGAUUUCAC_A_______ ......................((.((((((......).))))).)).......((((((....).))))).............................................. (-11.97 = -11.39 + -0.58)
| Location | 9,593,265 – 9,593,374 |
|---|---|
| Length | 109 |
| Sequences | 9 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 65.95 |
| Shannon entropy | 0.67359 |
| G+C content | 0.47532 |
| Mean single sequence MFE | -20.29 |
| Consensus MFE | -10.35 |
| Energy contribution | -10.48 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -0.78 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.810972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
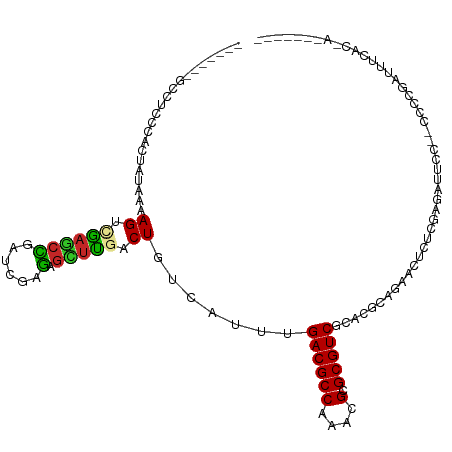
>dm3.chr3R 9593265 109 + 27905053 CUUCACUGUCAUUUGACGCCAAACGCGCGUCGCACGCAGAAGUCUCGAGAUUCCCCC-GAUUUCACAAUAUAGUAUAUAUAUAGUUAUAUAUUCAAUUCUUCGCCGGAUC----- .....(((......((((((....).)))))....((.((((....((((((.....-)))))).......((((((((......)))))))).....)))))))))...----- ( -20.30, z-score = -0.69, R) >droSim1.chr3R 15684274 109 - 27517382 CUCGACUGUCAUUUGACGCCAAACGCGCGUCGCACGCAGAAGUCUCGAGAUUCCCCCCGAUUUCACUAUAUAAUAUAUAUAUAGUUAUAUAUUCAAUUCUUCGCCG-AUC----- .(((..........((((((....).)))))....((.((((....((((((......))))))((((((((.....)))))))).............))))))))-)..----- ( -22.30, z-score = -1.55, R) >droSec1.super_0 8705995 109 - 21120651 CUCGACUGUCAUUUGACGCCAAACGCGCGUCGCACGCAGAAGUCUCGAGAUUCCCCCCGAUUUCACUAUAUAAUAUAUAUAUAGUUAUAUAUUCAAUUCUUCGCCG-AUC----- .(((..........((((((....).)))))....((.((((....((((((......))))))((((((((.....)))))))).............))))))))-)..----- ( -22.30, z-score = -1.55, R) >droYak2.chr3R 13901727 108 + 28832112 CUCGACUGUCAUUUGACGCCAAACGCGCGUCGCACGCAGAACUCUCGAGAUUCCCCC-GAUUUCACUAUAUUAUAUAUACAUAGCUAUAUAUUCAAUUCUUCGCCG-AUC----- (((((((((....(((((((....).))))))...)))).....))))).......(-((............(((((((......)))))))........)))...-...----- ( -17.95, z-score = -0.43, R) >droEre2.scaffold_4770 5706824 104 - 17746568 CUCGACUGUCAUUUGACGCCAAACGCGCGUCGCACGCAGAACUCUCGAGAUUCCCCC-GAUUUCACUAUAUA----AUAUAUAGCUAUAUAUUCAAUUCUUCGCCG-CUC----- ..............((((((....).)))))((.((.((((.....((((((.....-)))))).((((((.----..))))))............)))).))..)-)..----- ( -17.90, z-score = -0.46, R) >droAna3.scaffold_13340 17919111 97 + 23697760 UUUGACUGUCAUUUGACGCCAAACGCGCGUCGUACGCAGAUCGCUCGAGAAACCCUCCGAU-----------------AUAUAAAUAUAUAUUCGACUCAUCCCCG-AUCCAGCC ...((((((....(((((((....).))))))...)))).))(((.(((.....)))((((-----------------((((....))))).)))...........-....))). ( -19.60, z-score = -1.22, R) >dp4.chr2 10142132 96 + 30794189 UUUGACUGUCAUUUGACGCCAAACGGGCGUCGCAAGAAGAACGCACAACGCACAAUC-GUUGGCUCCAGUCCAGUCCCGUCCAGUUGUUCCCACACC------------------ ...(((((......((((((.....))))))...........((.(((((......)-))))))..)))))..........................------------------ ( -24.20, z-score = -0.41, R) >droPer1.super_0 1483867 96 - 11822988 UUUGACUGUCAUUUGACGCCAAACGAGCGUCGCAAGAGGAACGCACAACGCACAAUC-GUUGGCUCCAGUCCAGUCCCGUCCAGUUGUUCCCAUACC------------------ ...(((((......((((((....).)))))......(((..((.(((((......)-)))))))))....))))).....................------------------ ( -23.30, z-score = -0.62, R) >droWil1.scaffold_181130 11047343 86 + 16660200 UUUGACUGUCAUUUGACGCCAAACGAGCGUCGUACGCAGUAGUU----AAUCCACAUCGGUUU----------------CCCAGUAGUAGUCUCUCUUCUCCCGCC--------- ....(((((....(((((((....).))))))...)))))....----.........(((...----------------...((.((....))))......)))..--------- ( -14.80, z-score = -0.04, R) >consensus CUUGACUGUCAUUUGACGCCAAACGCGCGUCGCACGCAGAACUCUCGAGAUUCCCCC_GAUUUCACUAUAUAAUAUAUAUAUAGUUAUAUAUUCAAUUCUUCGCCG_AUC_____ .....((((....(((((((....).))))))...))))............................................................................ (-10.35 = -10.48 + 0.12)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:13:01 2011