| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,558,712 – 9,558,812 |
| Length | 100 |
| Max. P | 0.929260 |

| Location | 9,558,712 – 9,558,812 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 69.24 |
| Shannon entropy | 0.56532 |
| G+C content | 0.53477 |
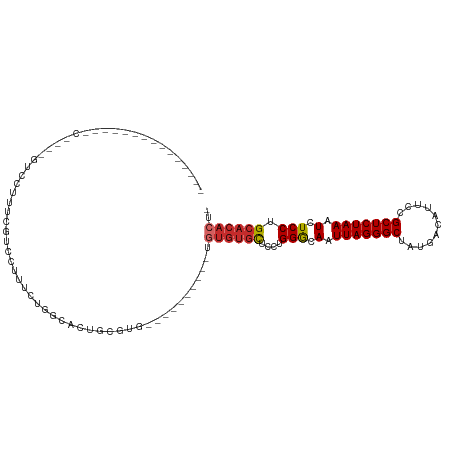
| Mean single sequence MFE | -25.52 |
| Consensus MFE | -15.17 |
| Energy contribution | -15.64 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.929260 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
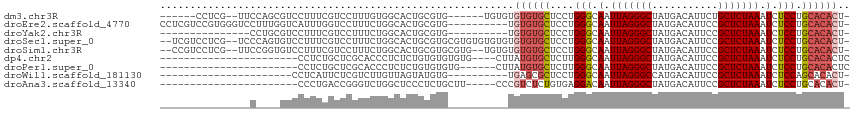
>dm3.chr3R 9558712 100 + 27905053 ------CCUCG--UUCCAGCGUCCUUUCGUCCUUUGUGGCACUGCGUG------UGUGUGUGUGCUCCUGGGCAAUUAGGGCUAUGACAUUCUGCUCUAAAUCUCCUGCACACU- ------.(.((--..((((.((((...........).))).))).)..------)).).((((((....(((.(.(((((((...........))))))).).))).)))))).- ( -24.50, z-score = 0.07, R) >droEre2.scaffold_4770 5671232 104 - 17746568 CCUCGUCCGUGGGUCCUUUGGUCAUUUGGUCCUUUCUGGCACUGCGUG----------UGUGUGCUCCUGGGCAAUUAGGGCUAUGACAUUCCGCUCUAAAUCUCCUGCACACU- ....(..((..((((....((.(.....).)).....))).)..))..----------)((((((....(((.(.(((((((...........))))))).).))).)))))).- ( -27.70, z-score = -0.02, R) >droYak2.chr3R 13867002 89 + 28832112 ---------------CCUGCGUCCUUUCGUCCUUUCUGGCACUGCGUG----------UGUGUGCUCCUGGGCAAUUAGGGCUAUGACAUUCCGCUCUAAAUCUCCUGCACACU- ---------------...((((((.............)).)).))(((----------(((.((((....)))).(((((((...........))))))).......)))))).- ( -22.32, z-score = -0.67, R) >droSec1.super_0 8671677 110 - 21120651 --UCGUCCUCG--UCCCAGUGUCCUUUCGUCCUUUCUGGCACUGCGUGCGUGUGUGUGUGUGUGCUCCUGGGCAAUUAGGGCUAUGACAUUCCGCUCUAAAUCUCCUGCACACU- --.((..(.((--..((((((.((.............))))))).)..)).)..))...((((((....(((.(.(((((((...........))))))).).))).)))))).- ( -32.12, z-score = -1.81, R) >droSim1.chr3R 15649407 108 - 27517382 --CCGUCCUCG--UUCCGGUGUCCUUUCGUCCUUUCUGGCACUGCGUGCGUG--UGUGUGUGUGCUCCUGGGCAAUUAGGGCUAUGACAUUCCGCUCUAAAUCUCCUGCACACU- --.((..(.((--(.((((((.((.............))))))).).))).)--..)).((((((....(((.(.(((((((...........))))))).).))).)))))).- ( -31.12, z-score = -1.44, R) >dp4.chr2 10108804 88 + 30794189 -----------------------CCUCUGCUCGCACCCUCUCUGUGUGUGUG----CUUAUGUGCUCUUGGGCAAUUAGGGCUAUGACAUUCCGCUCUAAAUCUCCUGCACACUC -----------------------.....((.(((((.(.....).))))).)----)...(((((....(((.(.(((((((...........))))))).).))).)))))... ( -24.70, z-score = -2.77, R) >droPer1.super_0 1450330 86 - 11822988 -----------------------CCUCUGCUCGCACCCUCUCUGUGUGUG------CUUAUGUGCUCUUGGGCAAUUAGGGCUAUGACAUUCCGCUCUAAAUCUCCUGCACACUC -----------------------.....((.(((((.......))))).)------)...(((((....(((.(.(((((((...........))))))).).))).)))))... ( -23.70, z-score = -2.76, R) >droWil1.scaffold_181130 11011173 82 + 16660200 ----------------------CCUCAUUCUCGUCUUGUUAGUAUGUG----------UGAGCGCUCCUGGGCAAUUAGGGCCAUGACAUUCCGCUCUAAAUCUCCAGCACACU- ----------------------.(((((...(((((....)).))).)----------)))).....(((((.(.(((((((...........))))))).).)))))......- ( -17.30, z-score = 0.24, R) >droAna3.scaffold_13340 13221865 86 - 23697760 -----------------------CCCUGACCGGGUCUGGCUCCCUCUGCUU-----CCCGUCUCUGUGAGGACAAUUAGGGCUAUGACAUUCCGCUCUAAAUCUCCUGCACACU- -----------------------....(((.(((...(((.......))).-----))))))..((((((((.(.(((((((...........))))))).).)))).))))..- ( -26.20, z-score = -2.11, R) >consensus _______________C____GUCCUUUCGUCCUUUCUGGCACUGCGUG__________UGUGUGCUCCUGGGCAAUUAGGGCUAUGACAUUCCGCUCUAAAUCUCCUGCACACU_ ...........................................................((((((....(((.(.(((((((...........))))))).).))).)))))).. (-15.17 = -15.64 + 0.47)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:12:59 2011