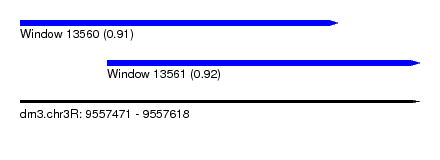
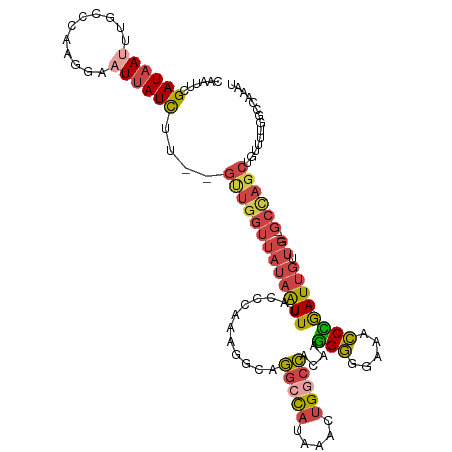
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,557,471 – 9,557,618 |
| Length | 147 |
| Max. P | 0.922719 |

| Location | 9,557,471 – 9,557,588 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 73.67 |
| Shannon entropy | 0.49862 |
| G+C content | 0.43380 |
| Mean single sequence MFE | -35.83 |
| Consensus MFE | -15.19 |
| Energy contribution | -16.67 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.909275 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9557471 117 + 27905053 CAAUUCGAUAAUUUGCCCAAGGCAUUAUCUU--GUUGGUUAUAAUUUACCCAAAGGCAGGCCAUAAACUGGCCACAAGGGGGAAACCCGAUUGUUGUUGGCCAGCUGUUUUGGCCAAAU .....(((((((((((((((((.....))))--)((((.((.....)).)))).))))(((((.....))))).....(((....)))))))))))((((((((.....)))))))).. ( -41.80, z-score = -2.46, R) >droAna3.scaffold_13340 13220998 88 - 23697760 AAACUCGAUAAUGCGCCCGACAAAUUAUCGG--CUUGAGGAUAGUUUAGCCAAGAAUUU-CUGCACAUAGUCGAUGCUGUUGCCAACAAAU---------------------------- ....(((((.(((.((.....(((((...((--((.((......)).))))...)))))-..)).))).))))).((....))........---------------------------- ( -15.70, z-score = 0.44, R) >droEre2.scaffold_4770 5670042 114 - 17746568 CAAUUCGAUAAUUUGUCCAAGAAUU-AUCUCUGGUUGGUUAUAAUUUACACAACGGCAGGACAUAAACUGGCCACAAGGGGGAA-CCCGAUUGUUG---GCUAGCUGUUUCGGCCAAAU ......((((((((......)))))-)))....((((((........)).))))(((.(((((....(((((((((.(((....-)))...)).))---))))).)))))..))).... ( -35.20, z-score = -2.72, R) >droYak2.chr3R 13865777 116 + 28832112 CAAUUCGAUAAUUUGCCCAAGGAUUUAUUUCUGGCUGGUUAUAAUUUACUCAAAGGCAGGCUAUAAACUGGCCACAAGGGGGAAACCCGAUUGUUG---GCCAGCUGUUUCGGCCAAAU .......(((((..(((..((((.....)))))))..)))))................((((..((.((((((((((.(((....)))..))).))---)))))...))..)))).... ( -38.80, z-score = -2.44, R) >droSec1.super_0 8670478 114 - 21120651 CAAUUCGAUAAUUUGCCCAAGGCAUUAUCUU--GUUGGUUAUAAUUUACACAAAGGCAGGCCAUAAACUGGCCACAUAGGGGAAACCUGAUUGUUG---GCCAGCUGUUUUGGCCAAAU ((((..(((((..((((...)))))))))..--)))).....................(((((.((.(((((((((..(((....)))...)).))---)))))...)).))))).... ( -40.70, z-score = -3.29, R) >droSim1.chr3R_random 547745 114 + 1307089 CAAUUCGAUAAUUUGCCCAAGGCAUUAUCUU--GUUGGUUAUAAUUUACCCAAAGGCAGGCCAUAAACUGGCCACAUGGGGGAAACCCGAUUGUUG---GCCAGCUGUUUUGGCCAAAU .............(((((((((.....))))--)((((.((.....)).)))).))))(((((.((.((((((((((.(((....))).).)).))---)))))...)).))))).... ( -42.80, z-score = -2.95, R) >consensus CAAUUCGAUAAUUUGCCCAAGGAAUUAUCUU__GUUGGUUAUAAUUUACCCAAAGGCAGGCCAUAAACUGGCCACAAGGGGGAAACCCGAUUGUUG___GCCAGCUGUUUUGGCCAAAU ......((((((...........))))))....((((((.((((((............(((((.....))))).....(((....))))))))).....)))))).............. (-15.19 = -16.67 + 1.48)

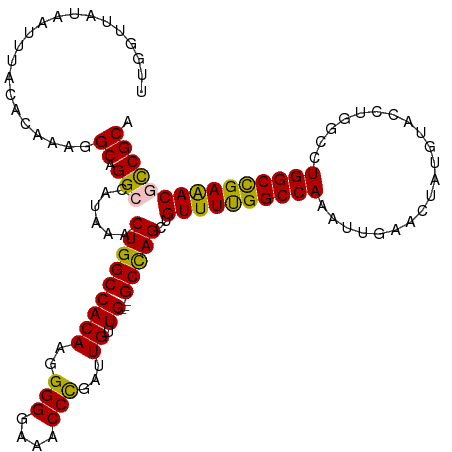
| Location | 9,557,503 – 9,557,618 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 90.96 |
| Shannon entropy | 0.15672 |
| G+C content | 0.49309 |
| Mean single sequence MFE | -41.37 |
| Consensus MFE | -36.02 |
| Energy contribution | -34.70 |
| Covariance contribution | -1.32 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.922719 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9557503 115 + 27905053 UUGGUUAUAAUUUACCCAAAGGCAGGCCAUAAACUGGCCACAAGGGGGAAACCCGAUUGUUGUUGGCCAGCUGUUUUGGCCAAAUUGA----UGUACCUUGCCUGGCCGAAACGCCGCA ((((.((.....)).))))..((.(((......((((((((((.(((....)))..))))....))))))..((((((((((......----...........))))))))))))))). ( -44.13, z-score = -2.14, R) >droEre2.scaffold_4770 5670075 115 - 17746568 UUGGUUAUAAUUUACACAACGGCAGGACAUAAACUGGCCACAAGGGGGAA-CCCGAUUGUUG---GCUAGCUGUUUCGGCCAAAUUGAACUAUGUACCUGGCCUGGCUGAGACAUCGCA .....................((..........(((((((((.(((....-)))...)).))---))))).(((((((((((.....................)))))))))))..)). ( -34.80, z-score = -1.11, R) >droYak2.chr3R 13865811 116 + 28832112 CUGGUUAUAAUUUACUCAAAGGCAGGCUAUAAACUGGCCACAAGGGGGAAACCCGAUUGUUG---GCCAGCUGUUUCGGCCAAAUUGAACUAUGUACCUUGCCUGGCUGAAACACCGCA .....................((.((.......((((((((((.(((....)))..))).))---))))).(((((((((((.....................))))))))))))))). ( -40.80, z-score = -2.44, R) >droSec1.super_0 8670510 116 - 21120651 UUGGUUAUAAUUUACACAAAGGCAGGCCAUAAACUGGCCACAUAGGGGAAACCUGAUUGUUG---GCCAGCUGUUUUGGCCAAAUUGAACUAUGUACCUGGCCUGGCCGAAACGCCGCA (((((........)).)))..((.(((......(((((((((..(((....)))...)).))---)))))..((((((((((.....................))))))))))))))). ( -41.80, z-score = -2.18, R) >droSim1.chr3R_random 547777 116 + 1307089 UUGGUUAUAAUUUACCCAAAGGCAGGCCAUAAACUGGCCACAUGGGGGAAACCCGAUUGUUG---GCCAGCUGUUUUGGCCAAAUUGAACUAUGUACCUGGCCUGGCCGAAACGCCGCA ((((.((.....)).))))..((.(((......((((((((((.(((....))).).)).))---)))))..((((((((((.....................))))))))))))))). ( -45.30, z-score = -2.38, R) >consensus UUGGUUAUAAUUUACACAAAGGCAGGCCAUAAACUGGCCACAAGGGGGAAACCCGAUUGUUG___GCCAGCUGUUUUGGCCAAAUUGAACUAUGUACCUGGCCUGGCCGAAACGCCGCA ..(((...............(((.(((((.....)))))(((..(((....)))...))).....)))....((((((((((.....................)))))))))))))... (-36.02 = -34.70 + -1.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:12:58 2011