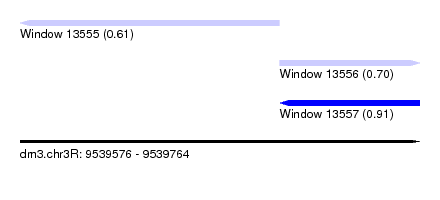
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,539,576 – 9,539,764 |
| Length | 188 |
| Max. P | 0.906996 |

| Location | 9,539,576 – 9,539,698 |
|---|---|
| Length | 122 |
| Sequences | 6 |
| Columns | 124 |
| Reading direction | reverse |
| Mean pairwise identity | 86.20 |
| Shannon entropy | 0.26348 |
| G+C content | 0.43758 |
| Mean single sequence MFE | -31.10 |
| Consensus MFE | -22.29 |
| Energy contribution | -23.18 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.606452 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
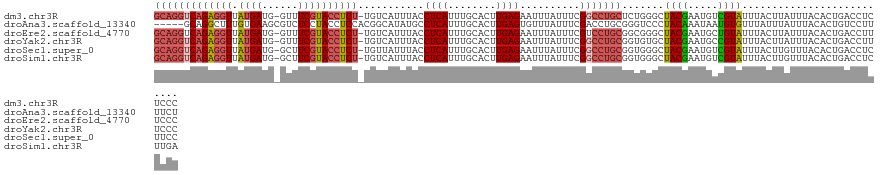
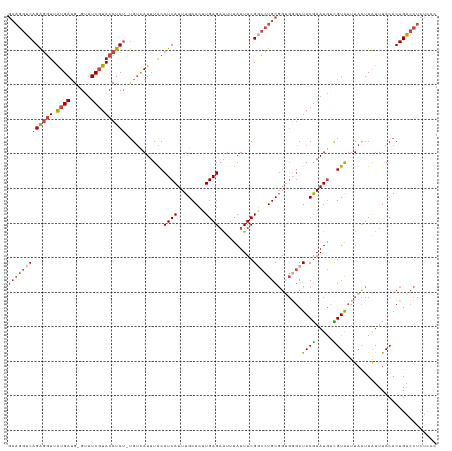
>dm3.chr3R 9539576 122 - 27905053 GCAGGUCAGAGGUUAUGAUG-GUUUCGUACCUCU-UGUCAUUUACCUCAUUUGCACUUGAGAAUUUAUUUCGGCCUGCUCUGGGCUACGAAUGUCGUAUUUACUUAUUUACACUGACCUCUCCC ..(((((((((((.((((..-...))))))))).-.......(((..((((((.......(((.....)))(((((.....))))).))))))..)))...............))))))..... ( -31.60, z-score = -2.07, R) >droAna3.scaffold_13340 13203557 119 + 23697760 -----GCAGGCUUUGUGAAGCGUCUCCUACCUCCACGGCAUAUGCCUCAUUUGCACUUGAGUGUUUAUUUCGACCUGCGGGUCCCUACAAAUAAUGUGUUUAUUUAUUUACACUGUCCUUUUCU -----(((((....(((.((.((.....)))).)))(((....)))...))))).....(((((..((...((((....)))).....((((((.....))))))))..))))).......... ( -22.20, z-score = 0.04, R) >droEre2.scaffold_4770 5650757 122 + 17746568 GCAGGUCAGAGGUUAUGAUG-GUUUCGUACCUCU-UGUCAUUUACCUCAUUUGCACUUGAGAAUUUAUUUCGUCCUGCGGCGGGCUACGAAUGCUGUAUUUACUUAUUUACACUGACCUUUCCC ..(((((((((((.((((((-((.....)))...-.)))))..)))))....(((.(((.(((.....)))((((......))))..))).)))...................))))))..... ( -30.80, z-score = -1.65, R) >droYak2.chr3R 13847962 122 - 28832112 GCAGGUCAGAGGUUAUGAUG-GUUUCGUACCUCU-UGUCAUUUACCUCAUUUGCACUUGAGAAUUUAUUUCGGCCUGCGGUGUGCUACGAAUGCCGUAUUUACUUAUUUACACUGACCUUUCCC (((((((((((((.((((..-...))))))))))-..........((((........))))..........)))))))((((((.((((.....))))..........)))))).......... ( -32.60, z-score = -2.33, R) >droSec1.super_0 8652638 122 + 21120651 GCAGGUCAGAGGUUAUGAUG-GCUUCGUACCUCU-UGUUAUUUACCUCAUUUGCACUUGAGAAUUUAUUUCGGCCUGCGGUGGGCUACGAAUGUCGUAUUUACUUGUUUACACUGACCUCUUCC (((((((((((((.((((..-...))))))))))-..........((((........))))..........)))))))(((.((.((((.....))))......((....)))).)))...... ( -34.70, z-score = -2.28, R) >droSim1.chr3R 15620850 122 + 27517382 GCAGGUCAGAGGUUAUGAUG-GCUUCGUACCUCU-UGUCAUUUACCUCAUUUGCACUUGAGAAUUUAUUUCGGCCUGCGGUGGGCUACGAAUGUCGUAUUUACUUGUUUACACUGACCUCUUGA (((((((((((((.((((..-...))))))))))-..........((((........))))..........)))))))(((.((.((((.....))))......((....)))).)))...... ( -34.70, z-score = -1.93, R) >consensus GCAGGUCAGAGGUUAUGAUG_GUUUCGUACCUCU_UGUCAUUUACCUCAUUUGCACUUGAGAAUUUAUUUCGGCCUGCGGUGGGCUACGAAUGUCGUAUUUACUUAUUUACACUGACCUCUCCC (((((((.(((((.((((......)))))))))............((((........))))..........))))))).......((((.....)))).......................... (-22.29 = -23.18 + 0.89)
| Location | 9,539,698 – 9,539,764 |
|---|---|
| Length | 66 |
| Sequences | 3 |
| Columns | 67 |
| Reading direction | forward |
| Mean pairwise identity | 75.68 |
| Shannon entropy | 0.31148 |
| G+C content | 0.59830 |
| Mean single sequence MFE | -7.77 |
| Consensus MFE | -6.71 |
| Energy contribution | -6.93 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.697651 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
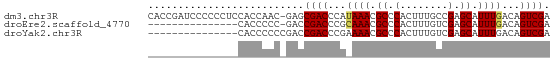
>dm3.chr3R 9539698 66 + 27905053 CACCGAUCCCCCCUCCACCAAC-GAGCGACCCAUAAACGCCCACUUUGCCGAGCAUUUGACAGUCGA ............(((.......-)))((((...((((.((.(........).)).))))...)))). ( -6.50, z-score = -0.53, R) >droEre2.scaffold_4770 5650879 51 - 17746568 ---------------CACCCCC-GACCGACCCGCAAACGCCCACUUUGUCGAGCAUUUGACAGUCGA ---------------.......-...((((...((((.((.(........).)).))))...)))). ( -9.30, z-score = -1.69, R) >droYak2.chr3R 13848084 52 + 28832112 ---------------CACCCCCCGACCGACCCGAAAACGCCCACUUUGUCGAGCAUUUGACAGUCGA ---------------...........((((.((....)).......((((((....)))))))))). ( -7.50, z-score = -1.01, R) >consensus _______________CACCCCC_GACCGACCCGAAAACGCCCACUUUGUCGAGCAUUUGACAGUCGA ..........................((((...((((.((.(........).)).))))...)))). ( -6.71 = -6.93 + 0.22)

| Location | 9,539,698 – 9,539,764 |
|---|---|
| Length | 66 |
| Sequences | 3 |
| Columns | 67 |
| Reading direction | reverse |
| Mean pairwise identity | 75.68 |
| Shannon entropy | 0.31148 |
| G+C content | 0.59830 |
| Mean single sequence MFE | -18.23 |
| Consensus MFE | -14.70 |
| Energy contribution | -14.70 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.906996 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
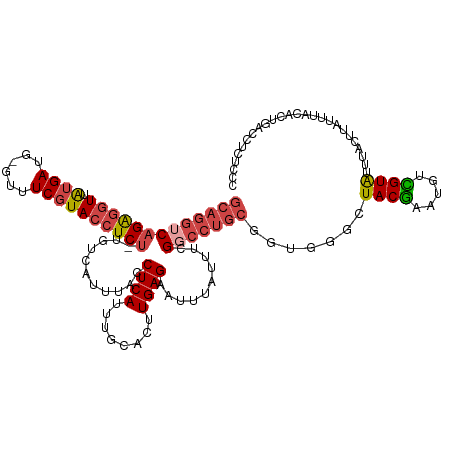
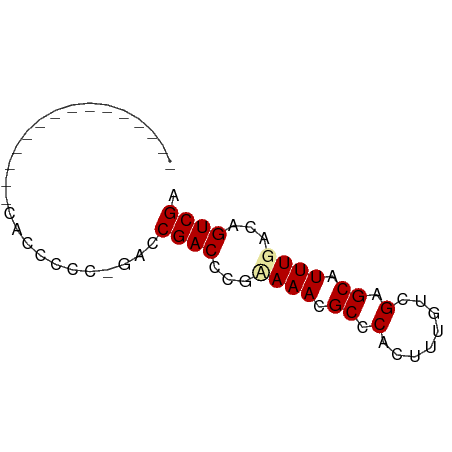
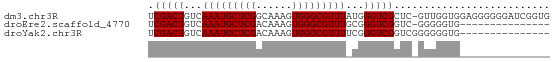
>dm3.chr3R 9539698 66 - 27905053 UCGACUGUCAAAUGCUCGGCAAAGUGGGCGUUUAUGGGUCGCUC-GUUGGUGGAGGGGGGAUCGGUG ...((((((...(.(((.((....((((((.........)))))-)...)).))).)..)).)))). ( -18.90, z-score = -0.84, R) >droEre2.scaffold_4770 5650879 51 + 17746568 UCGACUGUCAAAUGCUCGACAAAGUGGGCGUUUGCGGGUCGGUC-GGGGGUG--------------- (((((((.((((((((((......)))))))))).....)))))-)).....--------------- ( -19.00, z-score = -2.50, R) >droYak2.chr3R 13848084 52 - 28832112 UCGACUGUCAAAUGCUCGACAAAGUGGGCGUUUUCGGGUCGGUCGGGGGGUG--------------- (((((((.((((((((((......)))))))))....).)))))))......--------------- ( -16.80, z-score = -1.74, R) >consensus UCGACUGUCAAAUGCUCGACAAAGUGGGCGUUUACGGGUCGGUC_GGGGGUG_______________ .(((((...(((((((((......)))))))))...))))).......................... (-14.70 = -14.70 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:12:55 2011