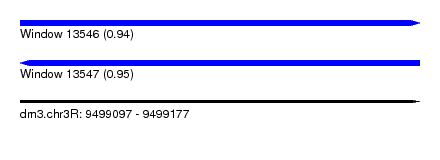
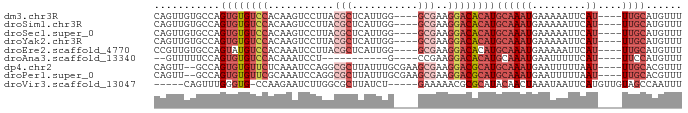
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,499,097 – 9,499,177 |
| Length | 80 |
| Max. P | 0.952559 |

| Location | 9,499,097 – 9,499,177 |
|---|---|
| Length | 80 |
| Sequences | 9 |
| Columns | 88 |
| Reading direction | forward |
| Mean pairwise identity | 76.11 |
| Shannon entropy | 0.47934 |
| G+C content | 0.42428 |
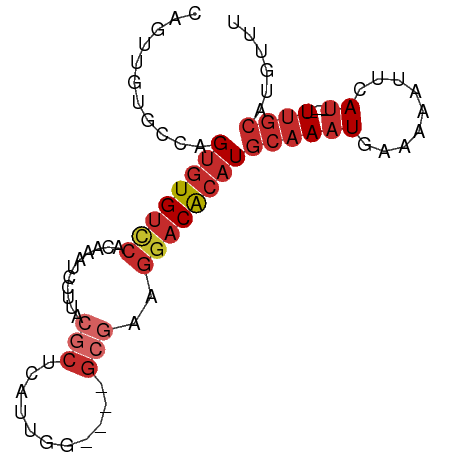
| Mean single sequence MFE | -24.17 |
| Consensus MFE | -8.18 |
| Energy contribution | -8.68 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.943577 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
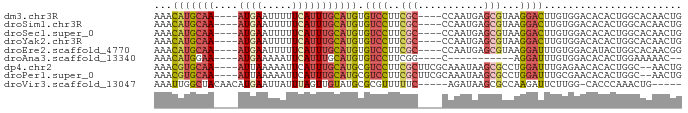
>dm3.chr3R 9499097 80 + 27905053 AAACAUGCAA----AUGAAUUUUUCAUUUGCAUGUGUCCUUCGC----CCAAUGAGCGUAAGGACUUGUGGACACACUGGCACAACUG ..((((((((----((((.....))))))))))))(((((((((----.......))).))))))(((((..(.....).)))))... ( -30.00, z-score = -4.55, R) >droSim1.chr3R 15580916 80 - 27517382 AAACAUGCAA----AUGAAUUUUUCAUUUGCAUGUGUCCUUCGC----CCAAUGAGCGUAAGGACUUGUGGACACACUGGCACAACUG ..((((((((----((((.....))))))))))))(((((((((----.......))).))))))(((((..(.....).)))))... ( -30.00, z-score = -4.55, R) >droSec1.super_0 8612709 80 - 21120651 AAACAUGCAA----AUGAAUUUUUCAUUUGCAUGUGUCCUUCGC----CCAAUGAGCGUAAGGACUUGUGGACACACUGGCACAACUG ..((((((((----((((.....))))))))))))(((((((((----.......))).))))))(((((..(.....).)))))... ( -30.00, z-score = -4.55, R) >droYak2.chr3R 13807447 80 + 28832112 AAACAUGCAA----AUGAAUUUUUCAUUUGCAUGUGUCCUUCGC----CCAAUGAGCGUAAGGACUUGUGGACACACUGGCACAACUG ..((((((((----((((.....))))))))))))(((((((((----.......))).))))))(((((..(.....).)))))... ( -30.00, z-score = -4.55, R) >droEre2.scaffold_4770 5610176 80 - 17746568 AAACAUGCAA----AUGAAUUUUUCAUUUGCAUGUGUCCUUCGC----CCAAUGAGCGUAAGGAUUUGUGGACAUACUGGCACAACGG ..((((((((----((((.....))))))))))))(((((((((----.......))).))))))(((((..(.....).)))))... ( -27.90, z-score = -3.61, R) >droAna3.scaffold_13340 2046363 67 + 23697760 AAACAUGGAA----AUGAAAAAUUCAUUUGCAUGUGUCCUUCGG----C-----------AGGAUUUGUGGACACACUGGAAAAAC-- ..(((((.((----((((.....)))))).)))))(((((....----.-----------)))))..(((....))).........-- ( -16.90, z-score = -2.00, R) >dp4.chr2 16940291 82 + 30794189 AAACGUGCAA----AUUAAAAAUUCAUUUGCAUGCGUCCUUCGCUUCGCAAAUAAGCGCCUGGAUUUGAGAACACACUGGC--AACUG ...(((((((----((.........))))))))).((((..(((((.......)))))...)))).............(..--..).. ( -16.70, z-score = -0.28, R) >droPer1.super_0 8373476 82 - 11822988 AAACGUGCAA----AUUAAAAAUUCAUUUGCAUGCGUCCUUCGCUUCGCAAAUAAGCGCCUGGAUUUGCGAACACACUGGC--AACUG ...(((((((----((.........))))))))).(((......(((((((((.((...))..)))))))))......)))--..... ( -19.10, z-score = -0.90, R) >droVir3.scaffold_13047 9272525 77 + 19223366 AAAUUGGCUACAACAUGAAUUAUUUAGUUGUAUGCGCGUUUUUC-----AGAUAAGCGCCAAGAUUCUUGG-CACCCAAACUG----- ...((((.((((((.(((.....)))))))))(((((((((...-----....))))))((((...)))))-)).))))....----- ( -16.90, z-score = -1.29, R) >consensus AAACAUGCAA____AUGAAUUUUUCAUUUGCAUGUGUCCUUCGC____CCAAUGAGCGUAAGGAUUUGUGGACACACUGGCACAACUG ...(((((((....((((.....))))))))))).((((..(((...........)))...))))....................... ( -8.18 = -8.68 + 0.50)
| Location | 9,499,097 – 9,499,177 |
|---|---|
| Length | 80 |
| Sequences | 9 |
| Columns | 88 |
| Reading direction | reverse |
| Mean pairwise identity | 76.11 |
| Shannon entropy | 0.47934 |
| G+C content | 0.42428 |
| Mean single sequence MFE | -26.36 |
| Consensus MFE | -7.83 |
| Energy contribution | -7.93 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.30 |
| Mean z-score | -3.21 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.952559 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
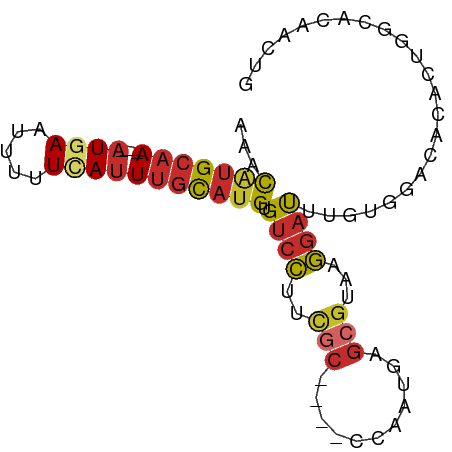
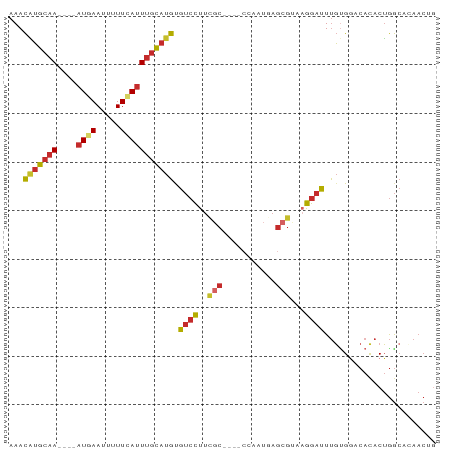
>dm3.chr3R 9499097 80 - 27905053 CAGUUGUGCCAGUGUGUCCACAAGUCCUUACGCUCAUUGG----GCGAAGGACACAUGCAAAUGAAAAAUUCAU----UUGCAUGUUU ...(((((.(.....)..)))))((((((.((((.....)----)))))))))((((((((((((.....))))----)))))))).. ( -32.50, z-score = -4.77, R) >droSim1.chr3R 15580916 80 + 27517382 CAGUUGUGCCAGUGUGUCCACAAGUCCUUACGCUCAUUGG----GCGAAGGACACAUGCAAAUGAAAAAUUCAU----UUGCAUGUUU ...(((((.(.....)..)))))((((((.((((.....)----)))))))))((((((((((((.....))))----)))))))).. ( -32.50, z-score = -4.77, R) >droSec1.super_0 8612709 80 + 21120651 CAGUUGUGCCAGUGUGUCCACAAGUCCUUACGCUCAUUGG----GCGAAGGACACAUGCAAAUGAAAAAUUCAU----UUGCAUGUUU ...(((((.(.....)..)))))((((((.((((.....)----)))))))))((((((((((((.....))))----)))))))).. ( -32.50, z-score = -4.77, R) >droYak2.chr3R 13807447 80 - 28832112 CAGUUGUGCCAGUGUGUCCACAAGUCCUUACGCUCAUUGG----GCGAAGGACACAUGCAAAUGAAAAAUUCAU----UUGCAUGUUU ...(((((.(.....)..)))))((((((.((((.....)----)))))))))((((((((((((.....))))----)))))))).. ( -32.50, z-score = -4.77, R) >droEre2.scaffold_4770 5610176 80 + 17746568 CCGUUGUGCCAGUAUGUCCACAAAUCCUUACGCUCAUUGG----GCGAAGGACACAUGCAAAUGAAAAAUUCAU----UUGCAUGUUU ...(((((.(.....)..))))).(((((.((((.....)----)))))))).((((((((((((.....))))----)))))))).. ( -28.70, z-score = -4.20, R) >droAna3.scaffold_13340 2046363 67 - 23697760 --GUUUUUCCAGUGUGUCCACAAAUCCU-----------G----CCGAAGGACACAUGCAAAUGAAUUUUUCAU----UUCCAUGUUU --((((((((((..(........)..))-----------)----..)))))))(((((.((((((.....))))----)).))))).. ( -15.70, z-score = -2.40, R) >dp4.chr2 16940291 82 - 30794189 CAGUU--GCCAGUGUGUUCUCAAAUCCAGGCGCUUAUUUGCGAAGCGAAGGACGCAUGCAAAUGAAUUUUUAAU----UUGCACGUUU .....--....(((((((((....((....(((......)))....)))))))))))((((((.........))----))))...... ( -19.10, z-score = 0.08, R) >droPer1.super_0 8373476 82 + 11822988 CAGUU--GCCAGUGUGUUCGCAAAUCCAGGCGCUUAUUUGCGAAGCGAAGGACGCAUGCAAAUGAAUUUUUAAU----UUGCACGUUU .....--....((((((((((........))((((.......))))...))))))))((((((.........))----))))...... ( -19.60, z-score = 0.35, R) >droVir3.scaffold_13047 9272525 77 - 19223366 -----CAGUUUGGGUG-CCAAGAAUCUUGGCGCUUAUCU-----GAAAAACGCGCAUACAACUAAAUAAUUCAUGUUGUAGCCAAUUU -----(((..((((((-(((((...))))))))))).))-----)......(.((.((((((............)))))))))..... ( -24.10, z-score = -3.69, R) >consensus CAGUUGUGCCAGUGUGUCCACAAAUCCUUACGCUCAUUGG____GCGAAGGACACAUGCAAAUGAAAAAUUCAU____UUGCAUGUUU .......((..((((((((..............................))))))))))............................. ( -7.83 = -7.93 + 0.10)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:12:46 2011