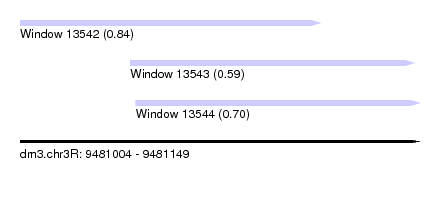
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,481,004 – 9,481,149 |
| Length | 145 |
| Max. P | 0.843113 |

| Location | 9,481,004 – 9,481,113 |
|---|---|
| Length | 109 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 69.21 |
| Shannon entropy | 0.63593 |
| G+C content | 0.58856 |
| Mean single sequence MFE | -40.26 |
| Consensus MFE | -21.65 |
| Energy contribution | -21.02 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.79 |
| Mean z-score | -0.87 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.843113 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9481004 109 + 27905053 GCCGAUGGCGGCAGGACGGAUUGGGCCAGAUAGGGAUUGUAGUUGGGAUUG---------UUGUAAUUGUAGCUAUCAGCAGUGGCCGGCUGGGGCGGAAUGUAGCCGGCUCGAACCG ((((....))))....(((.(((((((............(((((((.((((---------(((.............)))))))..))))))).(((........)))))))))).))) ( -39.32, z-score = -1.02, R) >dp4.chr2 16921828 106 + 30794189 GCCGAAGGCGGCAGGCUUGACUGUGCCAAAAACGGAUUGAAGU-GUGGAUU-----------GUAGCAGUUUCUGUCCAUCACGGCUGGCUGCGGCGGCACGUAGCCGGCGCGGGCCG ((((..((((.(((......)))))))......((((.(((((-((.....-----------...))).)))).))))....)))).(((((((.((((.....)))).))).)))). ( -47.50, z-score = -1.38, R) >droPer1.super_11055 717 106 - 964 GCUGAAGGCGGCAGGCUUGACUGUGCCAAAAACGGAUUGAAGU-GUGGAUU-----------GUAGCAGUUUCUGUCCAUCACGGCUGGCUGCGGUGGAACGUAGCCGGCGCGGGCCG ((((....)))).((((((.((((.......))))......((-((((((.-----------............))))).))).((((((((((......)))))))))).)))))). ( -44.12, z-score = -1.46, R) >droAna3.scaffold_13340 2029571 109 + 23697760 GCCGAUGGCGGGAGGACGGUCUGCGCCAGGUAGGGAUUGUAGGUGCUGCUG---------CUGUAGUUGUAGCUAUCCAGGGAGGCCGGUUGCGGCGGAAUGUAGCCGGCGCGGGCGG .(((....))).....(.(((((((((.(((..((((.((....(((((..---------..)))))....)).))))......)))(((((((......)))))))))))))))).) ( -45.60, z-score = -0.24, R) >droEre2.scaffold_4770 5591780 109 - 17746568 GCUGAUGGCGGCAGGACGGACUGGGCCAGGUAGGGAUUGUAGUUGGCGUUG---------UUGUAGUUGUAGCUAUCCAAAGUGGGCGGCUGGGGCGGAAUGUAGCCGGCCCGGAUCG ((((....))))....(((.(((((((.......(((....)))(((...(---------((...(((.(((((..((.....))..))))).)))..)))...)))))))))).))) ( -39.40, z-score = -0.31, R) >droYak2.chr3R 13789592 118 + 28832112 GCCGAUGGCGGCAGGACGGACUGGGCCAGGUAGGGAUUGUAGUUGGCAUUGCUGUUGUAGUUGUAGUUGUAGUUAUCCAAGGUGGGCGGCUGGGGCGGAAUGUAGCCGGCUCGGAUUG .((((.(.(((((.(((((.(...(((((.((.......)).)))))...)))))).).......(((.((((..(((.....)))..)))).)))........)))).))))).... ( -41.10, z-score = -0.81, R) >droSec1.super_0 8594431 109 - 21120651 GCCGAUGGCGGCAGAACGGACUGGGCCAGAUAGCGAUUGUAGUUGGGAUUG---------UUGUAGUUGUAGCUAUCAGCGGUGGCCGGCUGGGGCGGAAUGUAGCCGGCUCGGACCG ((((....))))....(((.(((((((............(((((((.((((---------(((((((....)))).)))))))..))))))).(((........)))))))))).))) ( -44.20, z-score = -1.59, R) >droSim1.chr3R 15562452 109 - 27517382 GCCGAUGGCGGCAGAACGGACUGGGCCAGAUAGGGAUGGUAGUUGGGAUUG---------UUGUAGUUGUAGCUAUCAGCGGUGGCCGGCUGGGGCGGAAUGUAGCCGGCUCGGACCG ((((....))))....(((.(((((((............(((((((.((((---------(((((((....)))).)))))))..))))))).(((........)))))))))).))) ( -44.20, z-score = -1.84, R) >droWil1.scaffold_181130 6565426 106 + 16660200 GCCGAUGGCGGCAAGGUGGUCUGCGCCAAAAACGGAUUAUAGCCAUGGCCA-----------AUUGUAAAAU-UAUCCAUCACUGCCGGCUGCGGUGGAAUGUAACCAGCACGAGCUG ((((.(((((((...((((((((.........)))))))).)))((((..(-----------(((....)))-)..))))....))))....))))..........((((....)))) ( -30.10, z-score = 1.04, R) >droVir3.scaffold_13047 9254383 106 + 19223366 GCCGAUGGUGGUAACGACGUUUGAGCCUGAUACGGAUUAUAGUUCGGAUUA------------UAGACUAAGUUAUCCAUAAUGGUCGGCUGGGGCGGAGCAUAACCGGCGCAUGCGG ((((((.((((((((...(((((....((((.(((((....))))).))))------------)))))...)))).))))....))))))((.(.(((.......))).).))..... ( -30.60, z-score = 0.04, R) >droMoj3.scaffold_6540 2769130 106 - 34148556 GCAGACGGCGGCAAUGAAGUUUGUGCCUGAUACGGAUUAUAAUUUGGAUUA------------UAGACUAAGUUGUCCAUGAUGGUCGGCUGUGGUGGCGCAUAGCCGGCGCAUGCGG (((..((.((((.........((((((...(((((.(((((((....))))------------))).)).(((((.((.....)).))))))))..))))))..)))).))..))).. ( -36.30, z-score = -1.16, R) >droGri2.scaffold_14906 10181151 106 - 14172833 GCCGAUGGUGGCAACGAGUGUUGCGCCUGAUACGGAUCAUAGUUUAGAUAG------------UAGACGGGAUCAUCCACAGUGGUUGGCUGUGGCGGUCCAUAGCCGGCACAAGCUG ...((((((.(((((....))))).((((.(((..(((........))).)------------))..))))))))))..((((.(((((((((((....)))))))))))....)))) ( -40.70, z-score = -1.77, R) >consensus GCCGAUGGCGGCAGGACGGACUGGGCCAGAUACGGAUUGUAGUUGGGAUUG___________GUAGUUGUAGCUAUCCAUGGUGGCCGGCUGGGGCGGAAUGUAGCCGGCGCGGGCCG ((((....))))....(((.(((((((.............................................((((.....))))..(((((.(......).)))))))))))).))) (-21.65 = -21.02 + -0.62)



| Location | 9,481,044 – 9,481,147 |
|---|---|
| Length | 103 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 63.13 |
| Shannon entropy | 0.75376 |
| G+C content | 0.58520 |
| Mean single sequence MFE | -39.60 |
| Consensus MFE | -16.26 |
| Energy contribution | -14.85 |
| Covariance contribution | -1.41 |
| Combinations/Pair | 2.00 |
| Mean z-score | -0.72 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.588218 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9481044 103 + 27905053 ---------AGUUGGGAUUGUUGUAAUUGUAGCUAUCAGCAGUGGCCGGCUGGGGCGGAAUGUAGCCGGCUCGAACCGAGCUAAAUAUGGGGAACCGAAAACGCCUGUCGGC------ ---------.(((((....((((......))))..)))))....((((((..(((((.....(((((((......))).))))....(((....)))....)))))))))))------ ( -35.80, z-score = -0.90, R) >dp4.chr2 16921868 102 + 30794189 ----------AGUGUGG--AUUGUAGCAGUUUCUGUCCAUCACGGCUGGCUGCGGCGGCACGUAGCCGGCGCGGGCCGCACCGCCCAUGGGAAAACGAAGGCGCUUGCCAGGAU---- ----------.((((((--((.............))))).)))(((.(((((((......)))))))(((((((((......)))).((......))...))))).))).....---- ( -41.32, z-score = 0.51, R) >droPer1.super_11055 757 102 - 964 ----------AGUGUGG--AUUGUAGCAGUUUCUGUCCAUCACGGCUGGCUGCGGUGGAACGUAGCCGGCGCGGGCCGCACCGCCCAUGGGAAAACGAAGGCGCUUGCCAGGAU---- ----------.((((((--((.............))))).)))(((.(((((((......)))))))(((((((((......)))).((......))...))))).))).....---- ( -41.32, z-score = -0.17, R) >droAna3.scaffold_13340 2029611 109 + 23697760 ---------AGGUGCUGCUGCUGUAGUUGUAGCUAUCCAGGGAGGCCGGUUGCGGCGGAAUGUAGCCGGCGCGGGCGGCUCCGCCCACGGGGAAGCGAAGACGUUUGCCGGGAUCUGG ---------....(((((.((....)).)))))...(((((...((((((((((......))))))))))..(((((....))))).(.((.(((((....))))).)).)..))))) ( -51.10, z-score = -0.95, R) >droEre2.scaffold_4770 5591820 109 - 17746568 ---------AGUUGGCGUUGUUGUAGUUGUAGCUAUCCAAAGUGGGCGGCUGGGGCGGAAUGUAGCCGGCCCGGAUCGAGCUGGAAACGGGGAAACGCAGACGCUUGCCGGGAUGGGG ---------.((((((...(((...(((.(((((..((.....))..))))).)))..)))...))))))((((..(((((.(....)(.(....).)....)))))))))....... ( -40.90, z-score = -0.80, R) >droYak2.chr3R 13789632 118 + 28832112 AGUUGGCAUUGCUGUUGUAGUUGUAGUUGUAGUUAUCCAAGGUGGGCGGCUGGGGCGGAAUGUAGCCGGCUCGGAUUGAACCGGAGACGGGGAACCGCAGACGCUUGCCGGGAUCGGG .....(((((((....)))).)))..........((((..((..((((.(((.((.....(((..((((.((.....)).))))..))).....)).))).))))..)).)))).... ( -39.70, z-score = 0.19, R) >droSec1.super_0 8594471 103 - 21120651 ---------AGUUGGGAUUGUUGUAGUUGUAGCUAUCAGCGGUGGCCGGCUGGGGCGGAAUGUAGCCGGCUCGGACCGAGCUGGAAAUGGGGAAGCGAAAACGCCUGUCGGG------ ---------.......(((((((((((....)))).)))))))..(((((..(((((........((((((((...))))))))......(....).....)))))))))).------ ( -36.40, z-score = -0.57, R) >droSim1.chr3R 15562492 103 - 27517382 ---------AGUUGGGAUUGUUGUAGUUGUAGCUAUCAGCGGUGGCCGGCUGGGGCGGAAUGUAGCCGGCUCGGACCGAGCUGGAAAUGGGGUAGCGAAAACGCCUGUCUGG------ ---------((((.(((((((((((((....)))).)))))))(((((((((.(......).)))))))))....)).))))(((...((.((.......)).))..)))..------ ( -37.30, z-score = -0.78, R) >droWil1.scaffold_181130 6565466 106 + 16660200 ---------AGCCAUGG---CCAAUUGUAAAAUUAUCCAUCACUGCCGGCUGCGGUGGAAUGUAACCAGCACGAGCUGCUCCACCCACAGGGAAACGUAAACGUUUGCCAGUGUCAUA ---------((((((((---..((((....))))..)))).......))))..((((((.......((((....)))).))))))(((.((.(((((....))))).)).)))..... ( -30.21, z-score = -0.29, R) >droVir3.scaffold_13047 9254423 106 + 19223366 ---------AGUUCGGA---UUAUAGACUAAGUUAUCCAUAAUGGUCGGCUGGGGCGGAGCAUAACCGGCGCAUGCGGCACCGCCAAUAGGUAGGCGUAUGCGUUUGCUGCAAUCCAA ---------.....(((---((.....((.((((..((.....))..)))).))..(.((((.....((((((((((.((((.......))).).)))))))))))))).)))))).. ( -37.00, z-score = -0.76, R) >droMoj3.scaffold_6540 2769170 106 - 34148556 ---------AAUUUGGA---UUAUAGACUAAGUUGUCCAUGAUGGUCGGCUGUGGUGGCGCAUAGCCGGCGCAUGCGGCGCCUCCAAUUGGCAAACGAACGCGUUUGCUGCAAUCCAG ---------....((((---((...(((......)))....(((((((((((((......)))))))))).)))((((((((.......)))(((((....)))))))))))))))). ( -43.50, z-score = -2.41, R) >droGri2.scaffold_14906 10181191 106 - 14172833 ---------AGUUUAGA---UAGUAGACGGGAUCAUCCACAGUGGUUGGCUGUGGCGGUCCAUAGCCGGCACAAGCUGCUCCGCCAAUGGGCAGGCGCAAACGUUUGCUGGUAUCAAA ---------........---(((((((((((.....))..(((.(((((((((((....)))))))))))....)))((.(((((....))).)).))...)))))))))........ ( -40.70, z-score = -1.69, R) >consensus _________AGUUGGGA__GUUGUAGUUGUAGCUAUCCAUGGUGGCCGGCUGGGGCGGAAUGUAGCCGGCGCGGGCCGCGCCGCCAAUGGGGAAACGAAAACGCUUGCCGGGAUC___ ...........................................(((.(((((((.(((.......))).)))).......(((....)))............))).)))......... (-16.26 = -14.85 + -1.41)


| Location | 9,481,046 – 9,481,149 |
|---|---|
| Length | 103 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 62.27 |
| Shannon entropy | 0.77186 |
| G+C content | 0.58567 |
| Mean single sequence MFE | -40.15 |
| Consensus MFE | -16.26 |
| Energy contribution | -14.85 |
| Covariance contribution | -1.41 |
| Combinations/Pair | 2.00 |
| Mean z-score | -0.79 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.704720 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9481046 103 + 27905053 ---------UUGGGAUUGUUGUAAUUGUAGCUAUCAGCAGUGGCCGGCUGGGGCGGAAUGUAGCCGGCUCGAACCGAGCUAAAUAUGGGGAACCGAAAACGCCUGUCGGCCU------ ---------.....(((((((.............)))))))(((((((..(((((.....(((((((......))).))))....(((....)))....)))))))))))).------ ( -39.12, z-score = -1.49, R) >dp4.chr2 16921870 100 + 30794189 ----------UGUGG--AUUGUAGCAGUUUCUGUCCAUCACGGCUGGCUGCGGCGGCACGUAGCCGGCGCGGGCCGCACCGCCCAUGGGAAAACGAAGGCGCUUGCCAGGAU------ ----------(.(((--...((.((.((((...(((((....((((((((((......))))))))))..((((......))))))))).))))....))))...))).)..------ ( -40.40, z-score = 0.60, R) >droPer1.super_11055 759 100 - 964 ----------UGUGG--AUUGUAGCAGUUUCUGUCCAUCACGGCUGGCUGCGGUGGAACGUAGCCGGCGCGGGCCGCACCGCCCAUGGGAAAACGAAGGCGCUUGCCAGGAU------ ----------(.(((--...((.((.((((...(((((....((((((((((......))))))))))..((((......))))))))).))))....))))...))).)..------ ( -40.40, z-score = -0.12, R) >droAna3.scaffold_13340 2029613 109 + 23697760 ---------GUGCUGCUGCUGUAGUUGUAGCUAUCCAGGGAGGCCGGUUGCGGCGGAAUGUAGCCGGCGCGGGCGGCUCCGCCCACGGGGAAGCGAAGACGUUUGCCGGGAUCUGGUU ---------..(((((.((....)).)))))...(((((...((((((((((......))))))))))..(((((....))))).(.((.(((((....))))).)).)..))))).. ( -51.70, z-score = -1.24, R) >droEre2.scaffold_4770 5591822 109 - 17746568 ---------UUGGCGUUGUUGUAGUUGUAGCUAUCCAAAGUGGGCGGCUGGGGCGGAAUGUAGCCGGCCCGGAUCGAGCUGGAAACGGGGAAACGCAGACGCUUGCCGGGAUGGGGCU ---------...(((((.(....(((.(((((..((.....))..))))).)))).)))))((((..(((((..(((((.(....)(.(....).)....))))))))))....)))) ( -41.40, z-score = -0.61, R) >droYak2.chr3R 13789634 118 + 28832112 UUGGCAUUGCUGUUGUAGUUGUAGUUGUAGUUAUCCAAGGUGGGCGGCUGGGGCGGAAUGUAGCCGGCUCGGAUUGAACCGGAGACGGGGAACCGCAGACGCUUGCCGGGAUCGGGCU ...(((((((....)))).)))......((((((((..((..((((.(((.((.....(((..((((.((.....)).))))..))).....)).))).))))..)).))))..)))) ( -40.90, z-score = 0.18, R) >droSec1.super_0 8594473 103 - 21120651 ---------UUGGGAUUGUUGUAGUUGUAGCUAUCAGCGGUGGCCGGCUGGGGCGGAAUGUAGCCGGCUCGGACCGAGCUGGAAAUGGGGAAGCGAAAACGCCUGUCGGGCU------ ---------.....(((((((((((....)))).)))))))(.(((((..(((((........((((((((...))))))))......(....).....)))))))))).).------ ( -38.20, z-score = -0.78, R) >droSim1.chr3R 15562494 103 - 27517382 ---------UUGGGAUUGUUGUAGUUGUAGCUAUCAGCGGUGGCCGGCUGGGGCGGAAUGUAGCCGGCUCGGACCGAGCUGGAAAUGGGGUAGCGAAAACGCCUGUCUGGCU------ ---------((((.(((((((((((....)))).)))))))(((((((((.(......).)))))))))....))))((..((...((.((.......)).))..))..)).------ ( -40.60, z-score = -1.43, R) >droWil1.scaffold_181130 6565468 106 + 16660200 ------------CCAUGGCCAAUUGUAAAAUUAUCCAUCACUGCCGGCUGCGGUGGAAUGUAACCAGCACGAGCUGCUCCACCCACAGGGAAACGUAAACGUUUGCCAGUGUCAUACU ------------..((((..((((....))))..))))((((......((.((((((.......((((....)))).))))))))..((.(((((....))))).))))))....... ( -27.60, z-score = 0.15, R) >droVir3.scaffold_13047 9254425 106 + 19223366 ------------UUCGGAUUAUAGACUAAGUUAUCCAUAAUGGUCGGCUGGGGCGGAGCAUAACCGGCGCAUGCGGCACCGCCAAUAGGUAGGCGUAUGCGUUUGCUGCAAUCCAACA ------------...(((((.....((.((((..((.....))..)))).))..(.((((.....((((((((((.((((.......))).).)))))))))))))).)))))).... ( -37.00, z-score = -0.94, R) >droMoj3.scaffold_6540 2769172 106 - 34148556 ------------UUUGGAUUAUAGACUAAGUUGUCCAUGAUGGUCGGCUGUGGUGGCGCAUAGCCGGCGCAUGCGGCGCCUCCAAUUGGCAAACGAACGCGUUUGCUGCAAUCCAGCA ------------.(((((((...(((......)))....(((((((((((((......)))))))))).)))((((((((.......)))(((((....))))))))))))))))).. ( -43.80, z-score = -1.93, R) >droGri2.scaffold_14906 10181193 106 - 14172833 ------------UUUAGAUAGUAGACGGGAUCAUCCACAGUGGUUGGCUGUGGCGGUCCAUAGCCGGCACAAGCUGCUCCGCCAAUGGGCAGGCGCAAACGUUUGCUGGUAUCAAACA ------------......(((((((((((.....))..(((.(((((((((((....)))))))))))....)))((.(((((....))).)).))...))))))))).......... ( -40.70, z-score = -1.82, R) >consensus __________UGUGGUUGUUGUAGUUGUAGCUAUCCAUGGUGGCCGGCUGGGGCGGAAUGUAGCCGGCGCGGGCCGCGCCGCCAAUGGGGAAACGAAAACGCUUGCCGGGAUC___C_ .........................................(((.(((((((.(((.......))).)))).......(((....)))............))).)))........... (-16.26 = -14.85 + -1.41)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:12:43 2011