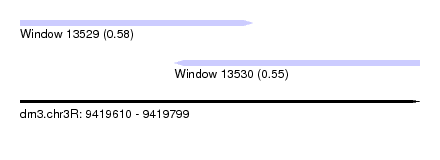
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,419,610 – 9,419,799 |
| Length | 189 |
| Max. P | 0.580465 |

| Location | 9,419,610 – 9,419,720 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 80.44 |
| Shannon entropy | 0.30265 |
| G+C content | 0.43120 |
| Mean single sequence MFE | -18.12 |
| Consensus MFE | -11.31 |
| Energy contribution | -11.31 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.580465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
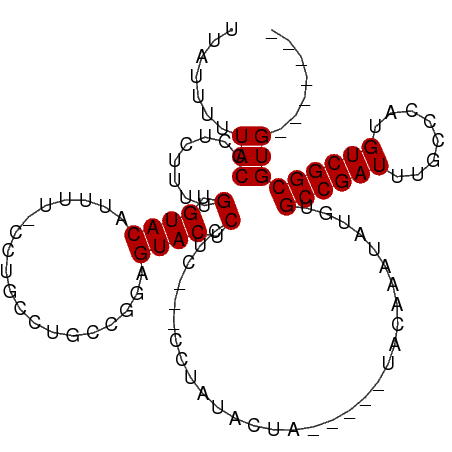
>dm3.chr3R 9419610 110 + 27905053 UUUCUCUUACCCUCUUUUGUGUACAAUGUUCCUGGCUGCCGUUGUACUCCUCACCCCUAUACUACAAUAUAUAAAUAUGUGCCGAUUUGCCCAUGUCGGCGUGAACAUAC .................(((((((((((...........))))))))...((((............(((((....)))))((((((........)))))))))))))... ( -21.00, z-score = -2.14, R) >droEre2.scaffold_4770 5524313 93 - 17746568 UUAUUUUUACCCUCUUUUGUGUACAUUUUGCCCGCCUGC--GAGUACUCUUC---CUCAUACUA-----UUCAAAUAUGUGCCGAUUUGCCCAUGUCGGCGUG------- ..................(.((((...((((......))--)))))).)...---.........-----...........((((((........))))))...------- ( -14.80, z-score = -1.27, R) >droYak2.chr3R 13728806 94 + 28832112 UUAUUUUUACCCUCUUUUGUGUACAUUUU-CCUGCCUGCCUGAGUACUCAUC---CCUAUACUA-----UUCAAAUAUGUGCCGAUUUUCCCAUGUCGGCGUG------- .................((.((((.....-.............)))).))..---.........-----...........((((((........))))))...------- ( -12.57, z-score = -1.51, R) >droSec1.super_0 8533034 104 - 21120651 UUUUUUUUACCCUCUUUUGUGUACAUUUU-CCUGGCUGACGGAGUACUCCUCACCCCUAUACUA-----UAUAAAUAUGUGCCGAUUUGCCCAUGUCGGCGUGUACAGAC .................(.(((((((...-...((.(((.(((....)))))).))........-----...........((((((........))))))))))))).). ( -24.10, z-score = -2.58, R) >consensus UUAUUUUUACCCUCUUUUGUGUACAUUUU_CCUGCCUGCCGGAGUACUCCUC___CCUAUACUA_____UACAAAUAUGUGCCGAUUUGCCCAUGUCGGCGUG_______ ..................(.((((...................)))).)...............................((((((........)))))).......... (-11.31 = -11.31 + -0.00)


| Location | 9,419,683 – 9,419,799 |
|---|---|
| Length | 116 |
| Sequences | 9 |
| Columns | 124 |
| Reading direction | reverse |
| Mean pairwise identity | 78.39 |
| Shannon entropy | 0.43152 |
| G+C content | 0.40048 |
| Mean single sequence MFE | -22.48 |
| Consensus MFE | -14.49 |
| Energy contribution | -14.95 |
| Covariance contribution | 0.46 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.554840 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9419683 116 - 27905053 CAACUAUUGUCUUCAUUUUUCUUGCAAAUCAUGCAAGGCAAUCUAAAAUGUUGACACAUUGGCAAAC----ACAUAUGUGGUUGUAU----GUUCACGCCGACAUGGGCAAAUCGGCACAUAUU ........(((..(((((((((((((.....)))))))......))))))..)))((((..((((.(----((....))).))))))----))....(((((..........)))))....... ( -26.50, z-score = -0.82, R) >droWil1.scaffold_181130 6496865 94 - 16660200 CAACUAUUGUCUUCAUUUUUCUUGCAAAUCAUGCAAGGCAAUCUAAAAUGUUGACACAUUGGCAAAC----ACAUGCAU------------AUACACAGUGACUAAGACA-------------- .......((((..(((((((((((((.....)))))))......))))))..))))((((((((...----...)))..------------.....))))).........-------------- ( -17.21, z-score = -0.81, R) >dp4.chr2 16857734 116 - 30794189 CAACUAUUGUCUUCAUUUUUCUUGCAAAUCAUGCAAGGCAAUCUAAAAUGUUGACACAUUGGCAAACA---GCACAAACAAGCGCACAGC-ACACACGCCGACACGGGCACAUCGACACU---- .......((((..(((((((((((((.....)))))))......))))))..))))..(((((.....---)).)))....((.....))-......(((......)))...........---- ( -22.80, z-score = -1.00, R) >droPer1.super_0 8288436 116 + 11822988 CAACUAUUGUCUUCAUUUUUCUUGCAAAUCAUGCAAGGCAAUCUAAAAUGUUGACACAUUGGCAAACA---GCACAAACAAGCGCACAGC-ACACACGCCGACACGGGCACAACGACACU---- .......((((........(((((((.....)))))))...........((((.....(((((.....---)).)))....((.....))-......(((......))).))))))))..---- ( -23.50, z-score = -1.30, R) >droAna3.scaffold_13340 1969721 100 - 23697760 CAACUAUUGUCUUCAUUUUUCUUGCAAAUCAUGCAAGGCAAUCUAAAAUGUUGACACAUUGGCAAACACACACGCCGACAGGGGCACAUCGACACAUUCC------------------------ ...(((.((((..(((((((((((((.....)))))))......))))))..))))...)))...........(((......)))...............------------------------ ( -18.90, z-score = -1.06, R) >droEre2.scaffold_4770 5524376 106 + 17746568 CAACUAUUGUCUUCAUUUUUCUUGCAAAUCAUGCAAGGCAAUCUAAAAUGUUGACACAUUGGCAAAC----ACAUAUGU--------------ACACGCCGACAUGGGCAAAUCGGCACAUAUU ...(((.((((..(((((((((((((.....)))))))......))))))..))))...))).....----..((((((--------------....(((((..........))))))))))). ( -22.70, z-score = -1.55, R) >droYak2.chr3R 13728870 106 - 28832112 CAACUAUUGUCUUCAUUUUUCUUGCAAAUCAUGCAAGGCAAUCUAAAAUGUUGACACAUUGGCAAAC----ACAUACGU--------------ACACGCCGACAUGGGAAAAUCGGCACAUAUU ...(((.((((..(((((((((((((.....)))))))......))))))..))))...))).....----........--------------....(((((..........)))))....... ( -21.10, z-score = -1.44, R) >droSec1.super_0 8533101 115 + 21120651 CAACUAUUGUCUUCAUUUUUCUUGCAAAUCAUGCAAGGCAAUCUAAAAUGUUGACACAUUGGCAAAC----ACAUAUGU-AUUGUCU----GUACACGCCGACAUGGGCAAAUCGGCACAUAUU .......((((..(((((((((((((.....)))))))......))))))..))))....(((((((----(....)))-.))))).----......(((((..........)))))....... ( -24.80, z-score = -1.04, R) >droSim1.chr2R 4623589 115 - 19596830 CAACUAUUGUCUUCAUUUUUCUUGCAAAUCAUGCAAGGCAAUCUAAAAUGUUGACACAUUGGCAAAC----ACAUAUGU-AUUGUCU----GAACACGCCGACAUGGGCAAAUCGGCACAUAUU .......((((..(((((((((((((.....)))))))......))))))..))))....(((((((----(....)))-.))))).----......(((((..........)))))....... ( -24.80, z-score = -1.16, R) >consensus CAACUAUUGUCUUCAUUUUUCUUGCAAAUCAUGCAAGGCAAUCUAAAAUGUUGACACAUUGGCAAAC____ACAUAUGU_A__G_A_____ACACACGCCGACAUGGGCAAAUCGGCACAUAUU .......((((..(((((((((((((.....)))))))......))))))..))))..(((((..................................)))))...................... (-14.49 = -14.95 + 0.46)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:12:32 2011