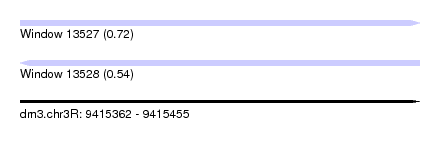
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,415,362 – 9,415,455 |
| Length | 93 |
| Max. P | 0.716592 |

| Location | 9,415,362 – 9,415,455 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 73.47 |
| Shannon entropy | 0.53368 |
| G+C content | 0.44362 |
| Mean single sequence MFE | -18.58 |
| Consensus MFE | -10.15 |
| Energy contribution | -10.67 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.716592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
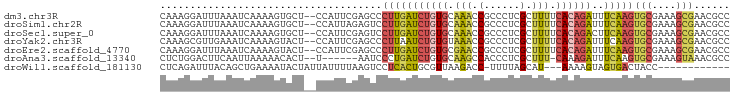
>dm3.chr3R 9415362 93 + 27905053 CAAAGGAUUUAAAUCAAAAGUGCU--CCAUUCGAGCCCUUGAUCUGUGCAAACCGCCCUCGCUUUUCACAGAUUUCAAGUGCGAAAGCGAACGCC .....................(((--(.....)))).(((((((((((.(((.((....)).))).))))))..)))))(((....)))...... ( -21.30, z-score = -1.13, R) >droSim1.chr2R 4619284 93 + 19596830 CAAAGGAUUUAAAUCAAAAGUGCU--CCAUUAGAGUCCUUGAUCUGUGCAAACCGCCCUCGCUUUUCACAGAUUUCAAGUGCGAAAGCGAACGCC ..((((((((.(((..........--..))).))))))))((((((((.(((.((....)).))).)))))))).....(((....)))...... ( -21.10, z-score = -1.29, R) >droSec1.super_0 8528805 93 - 21120651 CAAAGGAUUUAAAUCAAAAGUGCU--CCAUUCGAGUCCUUGAUCUGUGCAAACCGCCCUCGCUUUUCACAGACUUCAAGUGCGAAAGCGAACGCC ..(((((((..(((..........--..)))..)))))))..((((((.(((.((....)).))).)))))).......(((....)))...... ( -20.10, z-score = -0.70, R) >droYak2.chr3R 13724463 93 + 28832112 CAAAGCGUUGAAAUCAAAAGUACU--CCAUUCGAGCCCUUAAUCUGUGUAAACCGCCCUCGCUUUUCACAGAUUUCAAGUGCGAAAGCGAACGCC ....(((((((((((..(((..((--(.....)))..)))....((((.(((.((....)).))).))))))))))...(((....)))))))). ( -24.50, z-score = -3.38, R) >droEre2.scaffold_4770 5520052 93 - 17746568 CAAAGGAUUUAAAUCAAAAGUACU--CCAUUCGAGCCCUUGAUCUGUGCGAACCGCCCUCGCUUUUCACAGAUUUCAAGUGCGAAAGCGAACGCC ....(((................)--)).((((.((.(((((((((((.(((.((....)).))).))))))..))))).)).....)))).... ( -19.89, z-score = -0.83, R) >droAna3.scaffold_13340 1965342 86 + 23697760 CUCUGGACUUCAAUUAAAAACACU--U------AAUCCCUGAUCUGUGCAAGCCACCCUCGCUUU-CAAAGAUUUCAAGUGCGAAAGUAAACGCC ........................--.------.......(((((.((.((((.......)))).-)).))))).....(((....)))...... ( -11.00, z-score = 0.24, R) >droWil1.scaffold_181130 6485565 79 + 16660200 CUCAGAUUUACAGCUGAAAAUACUAUUAUUUUAAGUCCUCACUGCGUUAAGACC-UUUUAGCAU---AAAAGUAGUGACUACC------------ .((((........)))).................((..(((((((((((((...-.))))))..---....)))))))..)).------------ ( -12.20, z-score = -0.75, R) >consensus CAAAGGAUUUAAAUCAAAAGUACU__CCAUUCGAGUCCUUGAUCUGUGCAAACCGCCCUCGCUUUUCACAGAUUUCAAGUGCGAAAGCGAACGCC .....................................(((((((((((.(((.(......).))).))))))..)))))(((....)))...... (-10.15 = -10.67 + 0.52)


| Location | 9,415,362 – 9,415,455 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 73.47 |
| Shannon entropy | 0.53368 |
| G+C content | 0.44362 |
| Mean single sequence MFE | -23.74 |
| Consensus MFE | -14.25 |
| Energy contribution | -15.22 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.43 |
| Mean z-score | -0.74 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.543847 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
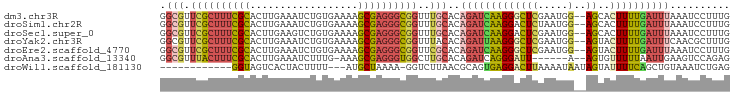
>dm3.chr3R 9415362 93 - 27905053 GGCGUUCGCUUUCGCACUUGAAAUCUGUGAAAAGCGAGGGCGGUUUGCACAGAUCAAGGGCUCGAAUGG--AGCACUUUUGAUUUAAAUCCUUUG .((.((((((((((((.........)))).)))))))).))((((((....((((((((((((.....)--))).).)))))))))))))..... ( -29.50, z-score = -1.70, R) >droSim1.chr2R 4619284 93 - 19596830 GGCGUUCGCUUUCGCACUUGAAAUCUGUGAAAAGCGAGGGCGGUUUGCACAGAUCAAGGACUCUAAUGG--AGCACUUUUGAUUUAAAUCCUUUG .((.((((((((((((.........)))).)))))))).))((((((....(((((((..(((.....)--))..))..)))))))))))..... ( -26.10, z-score = -1.28, R) >droSec1.super_0 8528805 93 + 21120651 GGCGUUCGCUUUCGCACUUGAAGUCUGUGAAAAGCGAGGGCGGUUUGCACAGAUCAAGGACUCGAAUGG--AGCACUUUUGAUUUAAAUCCUUUG .((.((((((((((((.........)))).)))))))).))((((((....(((((((..(((.....)--))..))..)))))))))))..... ( -25.90, z-score = -0.50, R) >droYak2.chr3R 13724463 93 - 28832112 GGCGUUCGCUUUCGCACUUGAAAUCUGUGAAAAGCGAGGGCGGUUUACACAGAUUAAGGGCUCGAAUGG--AGUACUUUUGAUUUCAACGCUUUG (((((.....((((..(((..((((((((.(((.((....)).))).))))))))..)))..))))(((--(((.(....))))))))))))... ( -26.10, z-score = -1.24, R) >droEre2.scaffold_4770 5520052 93 + 17746568 GGCGUUCGCUUUCGCACUUGAAAUCUGUGAAAAGCGAGGGCGGUUCGCACAGAUCAAGGGCUCGAAUGG--AGUACUUUUGAUUUAAAUCCUUUG .(((.((((((((((..................))))))))))..)))..(((((((((((((.....)--))..)))))))))).......... ( -28.37, z-score = -1.48, R) >droAna3.scaffold_13340 1965342 86 - 23697760 GGCGUUUACUUUCGCACUUGAAAUCUUUG-AAAGCGAGGGUGGCUUGCACAGAUCAGGGAUU------A--AGUGUUUUUAAUUGAAGUCCAGAG ((..((((.((..((((((((..((((((-(..(((((.....))))).....)))))))))------)--)))))....)).))))..)).... ( -19.90, z-score = -0.11, R) >droWil1.scaffold_181130 6485565 79 - 16660200 ------------GGUAGUCACUACUUUU---AUGCUAAAA-GGUCUUAACGCAGUGAGGACUUAAAAUAAUAGUAUUUUCAGCUGUAAAUCUGAG ------------(((..(((((.(((((---(...)))))-)((......)))))))..))).......(((((.......)))))......... ( -10.30, z-score = 1.14, R) >consensus GGCGUUCGCUUUCGCACUUGAAAUCUGUGAAAAGCGAGGGCGGUUUGCACAGAUCAAGGACUCGAAUGG__AGUACUUUUGAUUUAAAUCCUUUG .(((.((((((((((..................))))))))))..)))..((((((((((...............)))))))))).......... (-14.25 = -15.22 + 0.96)
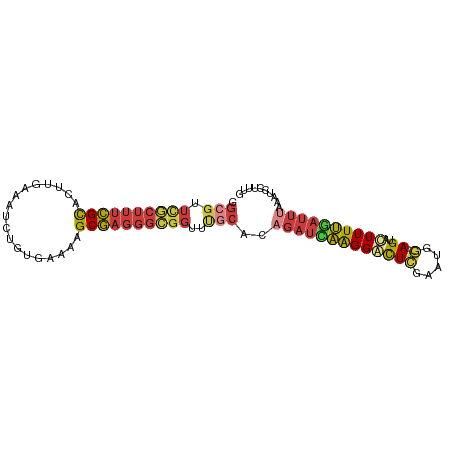
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:12:30 2011