
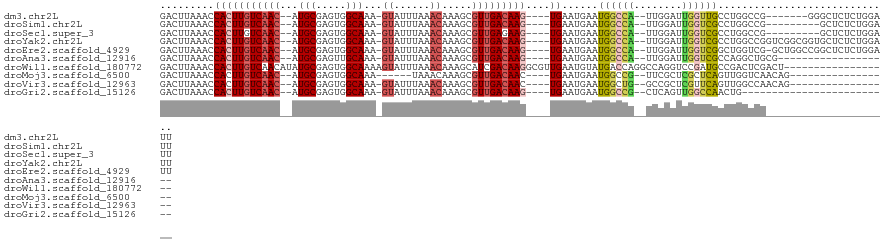
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,339,670 – 7,339,776 |
| Length | 106 |
| Max. P | 0.591270 |

| Location | 7,339,670 – 7,339,776 |
|---|---|
| Length | 106 |
| Sequences | 10 |
| Columns | 122 |
| Reading direction | reverse |
| Mean pairwise identity | 77.33 |
| Shannon entropy | 0.42980 |
| G+C content | 0.46256 |
| Mean single sequence MFE | -31.04 |
| Consensus MFE | -13.36 |
| Energy contribution | -13.11 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.591270 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 7339670 106 - 23011544 GACUUAAACCACUUGUCAAC--AUGCGAGUGGCAAA-GUAUUUAAACAAAGCGUUGACAAG----UGAAUGAAUGGCCA--UUGGAUUGGUUGCCUGGCCG-------GGGCUCUCUGGAUU (((((....(((((((((((--.(((.....)))..-((......)).....)))))))))----))......((((((--..((........))))))))-------))).))........ ( -29.70, z-score = -0.75, R) >droSim1.chr2L 7134857 104 - 22036055 GACUUAAACCACUUGUCAAC--AUGCGAGUGGCAAA-GUAUUUAAACAAAGCGUUGACAAG----UGAAUGAAUGGCCA--UUGGAUUGGUCGCCUGGCCG---------GCUCUCUGGAUU .........(((((((((((--.(((.....)))..-((......)).....)))))))))----)).........(((--..(..((((((....)))))---------)..)..)))... ( -30.70, z-score = -1.50, R) >droSec1.super_3 2861757 104 - 7220098 GACUUAAACCACUUGUCAAC--AUGCGAGUGGCAAA-GUAUUUAAACAAAGCGUUGAGAAG----UGAAUGAAUGGCCA--UUGGAUUGGUCGCCUGGCCG---------GCUCUCUGGAUU .((((...((((((((....--..))))))))..))-))................((((..----.........(((((--......)))))(((.....)---------))))))...... ( -27.00, z-score = -0.34, R) >droYak2.chr2L 16765503 113 + 22324452 GACUUAAACCACUUGUCAAC--AUGCGAGUGGCAAA-GUAUUUAAACAAAGCGUUGACAAG----UGAAUGAAUGGCCA--UUGGAUUGGUCGCCUGGCCGGUCGGCGGUGCUCUCUGGAUU .........(((((((((((--.(((.....)))..-((......)).....)))))))))----)).......(((((--......)))))((.(.(((....))).).)).......... ( -34.00, z-score = -0.83, R) >droEre2.scaffold_4929 16260997 112 - 26641161 GACUUAAACCACUUGUCAAC--AUGCGAGUGGCAAA-GUAUUUAAACAAAGCGUUGACAAG----UGAAUGAAUGGCCA--UUGGAUUGGUCGGCUGGUCG-GCUGGCCGGCUCUCUGGAUU .........(((((((((((--.(((.....)))..-((......)).....)))))))))----)).........(((--..(..((((((((((....)-)))))))))..)..)))... ( -39.80, z-score = -2.89, R) >droAna3.scaffold_12916 15248301 94 - 16180835 GACUUAAACCACUUGUCAAC--AUGCGAGUUGCAAA-GUAUUUAAACAAAGCGUUGACAAG----UGAAUGAAUGGCCA--UUGGAUUGGUCGCCAGGCUGCG------------------- .........(((((((((((--.(((.....)))..-((......)).....)))))))))----)).......(((((--......)))))((......)).------------------- ( -26.40, z-score = -1.42, R) >droWil1.scaffold_180772 3530476 104 + 8906247 GACUUAAACCACUUGUCAACAUAUGCGAGUGGCAAAAGUAUUUAAACAAAGCAUCGACAAGGCGUUGAAUGUAUGACCAGGCCAGGUCCGAUGCCGACUCGACU------------------ .((((...((((((((........))))))))...))))..............((((...(((((((.......((((......)))))))))))...))))..------------------ ( -29.91, z-score = -2.45, R) >droMoj3.scaffold_6500 13833963 91 + 32352404 GACUUAAACCACUUGUCAAC--AUGCGAGUGGCAAA------UAAACAAAGCGUUGACAAC----UGAAUGAAUGGCCG--UUCGCUCGCUCAGUUGGUCAACAG----------------- ........((((((((....--..))))))))....------..........(((((((((----(((.(((.(((...--.))).))).)))))).))))))..----------------- ( -28.10, z-score = -2.59, R) >droVir3.scaffold_12963 4679623 96 + 20206255 GACUUAAACCACUUGUCAAC--AUGCGAGUGGCAAA-GUAUUUAAACAAAGCGUUGACAAC----UGAAUGAAUGGCUG--GCCGCUCGUUCAGUUGGCCAACAG----------------- .((((...((((((((....--..))))))))..))-)).............((((.((((----(((((((.(((...--.))).)))))))))))..))))..----------------- ( -36.60, z-score = -4.56, R) >droGri2.scaffold_15126 4915659 88 - 8399593 GACUUAAACCACUUGUCAAC--AUGCGAGUGGCAAA-GUAUUUAAACAAAGCGUUGACAAG----UGAAUGAAUGGCCG--CUCAGUUGGCCAACUG------------------------- .........(((((((((((--.(((.....)))..-((......)).....)))))))))----))......((((((--......))))))....------------------------- ( -28.20, z-score = -3.01, R) >consensus GACUUAAACCACUUGUCAAC__AUGCGAGUGGCAAA_GUAUUUAAACAAAGCGUUGACAAG____UGAAUGAAUGGCCA__UUGGAUUGGUCGCCUGGCCG_____________________ ........((((((((........)))))))).........................................((((((........))))))............................. (-13.36 = -13.11 + -0.25)
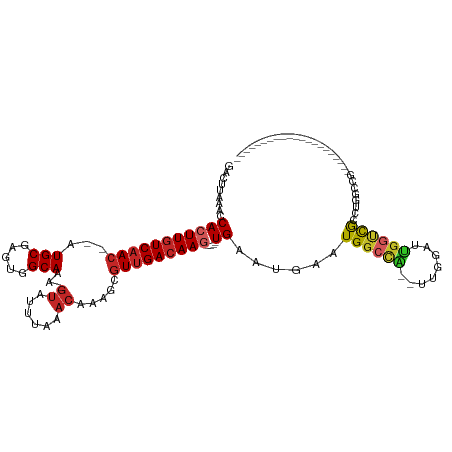
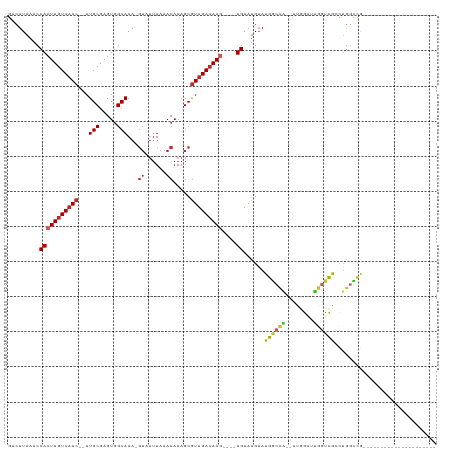
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:23:25 2011