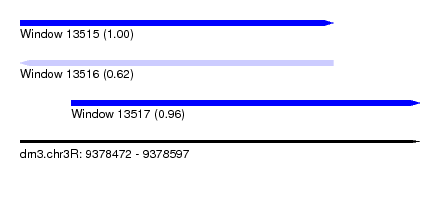
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,378,472 – 9,378,597 |
| Length | 125 |
| Max. P | 0.996915 |

| Location | 9,378,472 – 9,378,570 |
|---|---|
| Length | 98 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 81.29 |
| Shannon entropy | 0.36851 |
| G+C content | 0.33687 |
| Mean single sequence MFE | -22.08 |
| Consensus MFE | -17.50 |
| Energy contribution | -17.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.01 |
| SVM RNA-class probability | 0.996915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
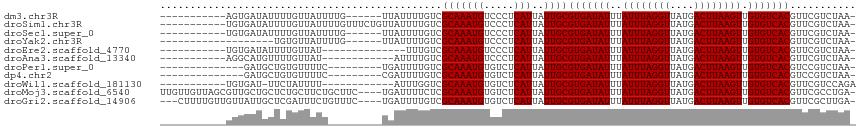
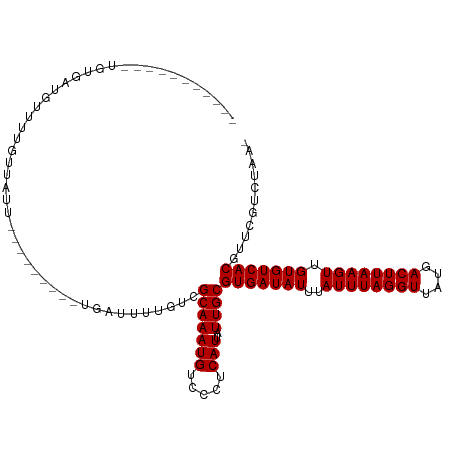
>dm3.chr3R 9378472 98 + 27905053 -----------AGUGAUAUUUUGUUAUUUUG------UUAUUUUGUCGCAAAUGUCCCUCAUUAUUGCGUGAUAUUUAUUUAGGUUAUGACUUAAGUUGUGUCACGUUCGUCUAA- -----------.((((((.............------......)))))).((((.....))))...(((((((((..((((((((....)))))))).)))))))))........- ( -21.51, z-score = -3.23, R) >droSim1.chr3R 15469997 104 - 27517382 -----------UGUGAUAUUUUGUUAUUUUGUUUCUGUUAUUUUGUCGCAAAUGUCCCUCAUUAUUGCGUGAUAUUUAUUUAGGUUAUGACUUAAGUUGUGUCACGUUCGUCUAA- -----------(((((((.........................)))))))((((.....))))...(((((((((..((((((((....)))))))).)))))))))........- ( -21.71, z-score = -3.24, R) >droSec1.super_0 8491893 98 - 21120651 -----------UGUGAUAUUUUGUUAUUUUG------UUAUUUUGUCGCAAAUGUCCCUCAUUAUUGCGUGAUAUUUAUUUAGGUUAUGACUUAAGUUGUGUCACGUUCGUCUAA- -----------(((((((.............------......)))))))((((.....))))...(((((((((..((((((((....)))))))).)))))))))........- ( -22.01, z-score = -3.40, R) >droYak2.chr3R 13685492 90 + 28832112 -------------------UGUGUUAUUUUG------UUAUUUUGUCGCAAAUGUCCCUCAUUAUUGCGUGAUAUUUAUUUAGGUUAUGACUUAAGUUGUGUCACGUUCGUCUAA- -------------------((((........------.........))))((((.....))))...(((((((((..((((((((....)))))))).)))))))))........- ( -17.73, z-score = -2.33, R) >droEre2.scaffold_4770 5482523 90 - 17746568 -----------UGUGAUAUUUUGUUAU--------------UUUGUCGCAAAUGUCCCUCAUUAUUGCGUGAUAUUUAUUUAGGUUAUGACUUAAGUUGUGUCACGUUCGUCUAA- -----------(((((((.........--------------..)))))))((((.....))))...(((((((((..((((((((....)))))))).)))))))))........- ( -22.60, z-score = -3.85, R) >droAna3.scaffold_13340 1931227 92 + 23697760 -----------AGGCAUGUUUUGUUAU------------AUUUUGUCGCAAAUGUCCCUCAUUAUUGCGUGAUAUUUAUUUAGGUUAUGACUUAAGUUGUGUCACGUUCGUCUAA- -----------((((....(((((.((------------.....)).)))))..............(((((((((..((((((((....)))))))).)))))))))..))))..- ( -21.50, z-score = -2.79, R) >droPer1.super_0 8250758 92 - 11822988 --------------GAUGCUGUGUUUUC---------UGAUUUUGUCGCAAAUGUGUCUCAUUAUUGCGUGAUAUUUAUUUAGGUUAUGACUUAAGUUGUGUCACGUCCGUCUAA- --------------((((..........---------.(((..((.(((....)))...))..)))(((((((((..((((((((....)))))))).))))))))).))))...- ( -21.90, z-score = -2.53, R) >dp4.chr2 16820659 92 + 30794189 --------------GAUGCUGUGUUUUC---------CGAUUUUGUCGCAAAUGUGUCUCAUUAUUGCGUGAUAUUUAUUUAGGUUAUGACUUAAGUUGUGUCACGUCCGUCUAA- --------------((((..........---------((((..((.(((....)))...))..))))((((((((..((((((((....)))))))).))))))))..))))...- ( -22.10, z-score = -2.51, R) >droWil1.scaffold_181130 6449918 92 + 16660200 -----------UGUGAU-UUCUAUUUU------------AUUUGGUCGCAAAUGUGUCUCAUUAUUGCGUGAUAUUUAUUUAGGUUAUGACUUAAGUUGUGUCACGUUCGUCCAGA -----------......-.........------------.(((((.((..((((.....))))...(((((((((..((((((((....)))))))).))))))))).)).))))) ( -23.10, z-score = -2.95, R) >droMoj3.scaffold_6540 2674230 111 - 34148556 UUGUUGUUAGCGUUGCUGCUCUGCUUCUGCUUC----UGAUUUUCUCGCAAAUGUGUCUCAUUAUUGCGUGAUAUUUAUUUAGGUUAUGACUUAAGUUGUGUCACGUUCGCCUGA- ..((((..((((..((......))...))))..----))))..((..((.((((.....))))...(((((((((..((((((((....)))))))).)))))))))..))..))- ( -24.50, z-score = -1.22, R) >droGri2.scaffold_14906 10093475 108 - 14172833 ---CUUUUGUUGUUAUUGCUCGAUUUCUGUUUC----UGAUUUUGUCGCAAAUGUGUCUCAUUAUUGCGUGAUAUUUAUUUAGGUUAUGACUUAAGUUGUGUCACGUUCGCUUGA- ---............((((.(((..((......----.))..)))..)))).......(((.....(((((((((..((((((((....)))))))).))))))))).....)))- ( -24.20, z-score = -2.62, R) >consensus ___________UGUGAUGUUUUGUUAUU_________UGAUUUUGUCGCAAAUGUCCCUCAUUAUUGCGUGAUAUUUAUUUAGGUUAUGACUUAAGUUGUGUCACGUUCGUCUAA_ ...............................................(((((((.....)))..))))(((((((..((((((((....)))))))).)))))))........... (-17.50 = -17.50 + 0.00)
| Location | 9,378,472 – 9,378,570 |
|---|---|
| Length | 98 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 81.29 |
| Shannon entropy | 0.36851 |
| G+C content | 0.33687 |
| Mean single sequence MFE | -10.71 |
| Consensus MFE | -9.00 |
| Energy contribution | -9.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.85 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.621460 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9378472 98 - 27905053 -UUAGACGAACGUGACACAACUUAAGUCAUAACCUAAAUAAAUAUCACGCAAUAAUGAGGGACAUUUGCGACAAAAUAA------CAAAAUAACAAAAUAUCACU----------- -......((..(((((.........))))).................(((((..(((.....)))))))).........------...............))...----------- ( -9.40, z-score = -0.79, R) >droSim1.chr3R 15469997 104 + 27517382 -UUAGACGAACGUGACACAACUUAAGUCAUAACCUAAAUAAAUAUCACGCAAUAAUGAGGGACAUUUGCGACAAAAUAACAGAAACAAAAUAACAAAAUAUCACA----------- -......((..(((((.........))))).................(((((..(((.....))))))))..............................))...----------- ( -9.40, z-score = -0.74, R) >droSec1.super_0 8491893 98 + 21120651 -UUAGACGAACGUGACACAACUUAAGUCAUAACCUAAAUAAAUAUCACGCAAUAAUGAGGGACAUUUGCGACAAAAUAA------CAAAAUAACAAAAUAUCACA----------- -......((..(((((.........))))).................(((((..(((.....)))))))).........------...............))...----------- ( -9.40, z-score = -0.72, R) >droYak2.chr3R 13685492 90 - 28832112 -UUAGACGAACGUGACACAACUUAAGUCAUAACCUAAAUAAAUAUCACGCAAUAAUGAGGGACAUUUGCGACAAAAUAA------CAAAAUAACACA------------------- -..........(((((.........))))).................(((((..(((.....)))))))).........------............------------------- ( -9.00, z-score = -0.89, R) >droEre2.scaffold_4770 5482523 90 + 17746568 -UUAGACGAACGUGACACAACUUAAGUCAUAACCUAAAUAAAUAUCACGCAAUAAUGAGGGACAUUUGCGACAAA--------------AUAACAAAAUAUCACA----------- -......((..(((((.........))))).................(((((..(((.....)))))))).....--------------...........))...----------- ( -9.40, z-score = -0.78, R) >droAna3.scaffold_13340 1931227 92 - 23697760 -UUAGACGAACGUGACACAACUUAAGUCAUAACCUAAAUAAAUAUCACGCAAUAAUGAGGGACAUUUGCGACAAAAU------------AUAACAAAACAUGCCU----------- -..........(((((.........))))).................(((((..(((.....)))))))).......------------................----------- ( -9.00, z-score = -0.18, R) >droPer1.super_0 8250758 92 + 11822988 -UUAGACGGACGUGACACAACUUAAGUCAUAACCUAAAUAAAUAUCACGCAAUAAUGAGACACAUUUGCGACAAAAUCA---------GAAAACACAGCAUC-------------- -......((..(((((.........)))))..)).............(((((..(((.....)))))))).........---------..............-------------- ( -11.20, z-score = -1.20, R) >dp4.chr2 16820659 92 - 30794189 -UUAGACGGACGUGACACAACUUAAGUCAUAACCUAAAUAAAUAUCACGCAAUAAUGAGACACAUUUGCGACAAAAUCG---------GAAAACACAGCAUC-------------- -......((..(((((.........)))))..)).............(((((..(((.....)))))))).........---------..............-------------- ( -11.20, z-score = -0.74, R) >droWil1.scaffold_181130 6449918 92 - 16660200 UCUGGACGAACGUGACACAACUUAAGUCAUAACCUAAAUAAAUAUCACGCAAUAAUGAGACACAUUUGCGACCAAAU------------AAAAUAGAA-AUCACA----------- ..(((......(((((.........))))).................(((((..(((.....)))))))).)))...------------.........-......----------- ( -11.80, z-score = -1.50, R) >droMoj3.scaffold_6540 2674230 111 + 34148556 -UCAGGCGAACGUGACACAACUUAAGUCAUAACCUAAAUAAAUAUCACGCAAUAAUGAGACACAUUUGCGAGAAAAUCA----GAAGCAGAAGCAGAGCAGCAACGCUAACAACAA -...((((...(((((.........)))))..............((.(((((..(((.....)))))))).))......----.........((......))..))))........ ( -16.40, z-score = -0.66, R) >droGri2.scaffold_14906 10093475 108 + 14172833 -UCAAGCGAACGUGACACAACUUAAGUCAUAACCUAAAUAAAUAUCACGCAAUAAUGAGACACAUUUGCGACAAAAUCA----GAAACAGAAAUCGAGCAAUAACAACAAAAG--- -....((....(((((.........))))).................(((((..(((.....)))))))).........----..............))..............--- ( -11.60, z-score = -1.09, R) >consensus _UUAGACGAACGUGACACAACUUAAGUCAUAACCUAAAUAAAUAUCACGCAAUAAUGAGGGACAUUUGCGACAAAAUAA_________AAUAACAAAACAUCACA___________ ...........(((((.........))))).................(((((..(((.....)))))))).............................................. ( -9.00 = -9.00 + 0.00)

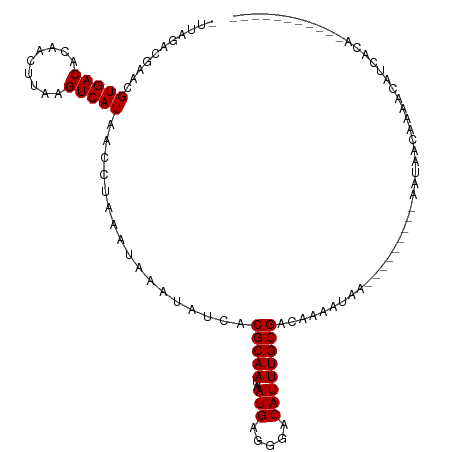
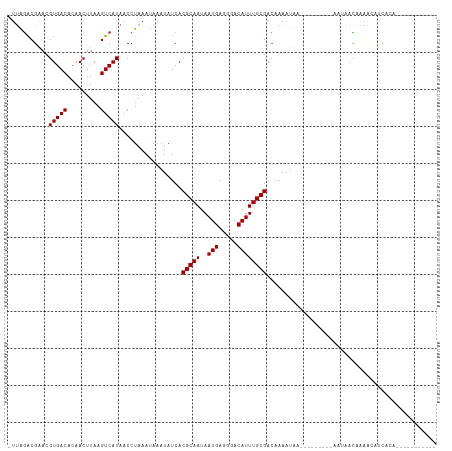
| Location | 9,378,488 – 9,378,597 |
|---|---|
| Length | 109 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 91.81 |
| Shannon entropy | 0.16561 |
| G+C content | 0.32600 |
| Mean single sequence MFE | -22.49 |
| Consensus MFE | -20.50 |
| Energy contribution | -20.32 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.961775 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9378488 109 + 27905053 ------UUUGUUAUUUUGUCGCAAAUGUCCCUCAUUAUUGCGUGAUAUUUAUUUAGGUUAUGACUUAAGUUGUGUCACGUUCGUCUA-AAUUUAUAUCCAAUA--UGUUGACUAUCAG ------...........((((((((((.....))))...(((((((((..((((((((....)))))))).))))))))).......-...............--)).))))...... ( -21.00, z-score = -2.09, R) >droSim1.chr3R 15470013 115 - 27517382 UUUGUUUCUGUUAUUUUGUCGCAAAUGUCCCUCAUUAUUGCGUGAUAUUUAUUUAGGUUAUGACUUAAGUUGUGUCACGUUCGUCUA-AAUUUAUAUCCAAUA--UGUUGACUAUCAG .................((((((((((.....))))...(((((((((..((((((((....)))))))).))))))))).......-...............--)).))))...... ( -21.00, z-score = -1.92, R) >droSec1.super_0 8491909 109 - 21120651 ------UUUGUUAUUUUGUCGCAAAUGUCCCUCAUUAUUGCGUGAUAUUUAUUUAGGUUAUGACUUAAGUUGUGUCACGUUCGUCUA-AAUUUAUAUCCAAUA--UGUUGACUAUCAG ------...........((((((((((.....))))...(((((((((..((((((((....)))))))).))))))))).......-...............--)).))))...... ( -21.00, z-score = -2.09, R) >droYak2.chr3R 13685502 107 + 28832112 --------UGUUAUUUUGUCGCAAAUGUCCCUCAUUAUUGCGUGAUAUUUAUUUAGGUUAUGACUUAAGUUGUGUCACGUUCGUCUA-AAUUUAUAUCCAAUA--UGUUGACUAUCAG --------.........((((((((((.....))))...(((((((((..((((((((....)))))))).))))))))).......-...............--)).))))...... ( -21.00, z-score = -2.03, R) >droEre2.scaffold_4770 5482533 107 - 17746568 --------UGUUAUUUUGUCGCAAAUGUCCCUCAUUAUUGCGUGAUAUUUAUUUAGGUUAUGACUUAAGUUGUGUCACGUUCGUCUA-AAUUUAUAUCCAAUA--UGUUGACUAUCAG --------.........((((((((((.....))))...(((((((((..((((((((....)))))))).))))))))).......-...............--)).))))...... ( -21.00, z-score = -2.03, R) >droAna3.scaffold_13340 1931237 109 + 23697760 ------UGUUAUAUUUUGUCGCAAAUGUCCCUCAUUAUUGCGUGAUAUUUAUUUAGGUUAUGACUUAAGUUGUGUCACGUUCGUCUA-AAUUUAUAUCCAAUA--UGUUGACUAUCAG ------...........((((((((((.....))))...(((((((((..((((((((....)))))))).))))))))).......-...............--)).))))...... ( -21.00, z-score = -1.86, R) >droPer1.super_0 8250770 107 - 11822988 --------UCUGAUUUUGUCGCAAAUGUGUCUCAUUAUUGCGUGAUAUUUAUUUAGGUUAUGACUUAAGUUGUGUCACGUCCGUCUA-AAUUUAUAUCCAAUA--UGUUGACUAUCAG --------.(((((...((((((((((.....))))...(((((((((..((((((((....)))))))).))))))))).......-...............--)).)))).))))) ( -25.60, z-score = -3.12, R) >dp4.chr2 16820671 107 + 30794189 --------UCCGAUUUUGUCGCAAAUGUGUCUCAUUAUUGCGUGAUAUUUAUUUAGGUUAUGACUUAAGUUGUGUCACGUCCGUCUA-AAUUUAUAUCCAAUA--UGUUGACUAUCAG --------...(((...((((((((((.....))))...(((((((((..((((((((....)))))))).))))))))).......-...............--)).)))).))).. ( -23.30, z-score = -2.35, R) >droWil1.scaffold_181130 6449929 108 + 16660200 --------UUUUAUUUGGUCGCAAAUGUGUCUCAUUAUUGCGUGAUAUUUAUUUAGGUUAUGACUUAAGUUGUGUCACGUUCGUCCAGAAUUUAUAUCCAAUA--UGUUGACUAUCAG --------.....(((((.((..((((.....))))...(((((((((..((((((((....)))))))).))))))))).)).)))))..............--............. ( -23.90, z-score = -2.11, R) >droVir3.scaffold_13047 9166252 107 + 19223366 ------UUUGUGAUUUUGUCGCAAAUGUGUCUCAUUAUUGCGUGAUAUUUAUUUAGGUUAUGACUUAAGUUGUGUCACGUUCGCCUG-AAUUUAUAUCCAAUA--UGUUGGCCAUC-- ------.(((((((...)))))))(((.(((....(((((...(((((..((((((((..((((.........)))).....)))))-)))..))))))))))--....)))))).-- ( -26.40, z-score = -2.56, R) >droMoj3.scaffold_6540 2674259 109 - 34148556 ------CUUCUGAUUUUCUCGCAAAUGUGUCUCAUUAUUGCGUGAUAUUUAUUUAGGUUAUGACUUAAGUUGUGUCACGUUCGCCUG-AAUUUAUAUCCAAUAUAUGCUGGCCAUC-- ------..................(((.(((.((((((((...(((((..((((((((..((((.........)))).....)))))-)))..)))))))))).)))..)))))).-- ( -21.80, z-score = -1.24, R) >droGri2.scaffold_14906 10093501 107 - 14172833 ------UUUCUGAUUUUGUCGCAAAUGUGUCUCAUUAUUGCGUGAUAUUUAUUUAGGUUAUGACUUAAGUUGUGUCACGUUCGCUUG-AAUUUAUAUCCAAUA--UGUUGGCCAUC-- ------...........(.(((....))).)(((.....(((((((((..((((((((....)))))))).))))))))).....))-)........((((..--..)))).....-- ( -22.90, z-score = -1.62, R) >consensus ______U_UGUUAUUUUGUCGCAAAUGUCCCUCAUUAUUGCGUGAUAUUUAUUUAGGUUAUGACUUAAGUUGUGUCACGUUCGUCUA_AAUUUAUAUCCAAUA__UGUUGACUAUCAG .................((((((((((.....))))...(((((((((..((((((((....)))))))).))))))))).........................))).)))...... (-20.50 = -20.32 + -0.18)

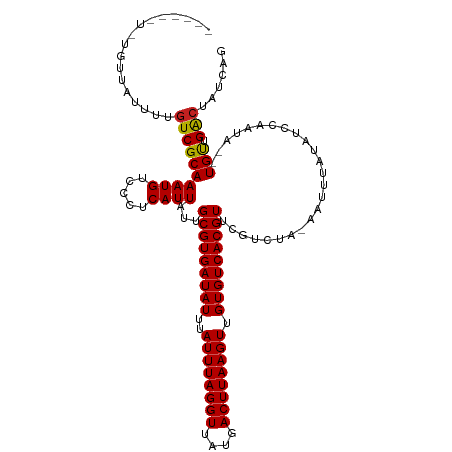
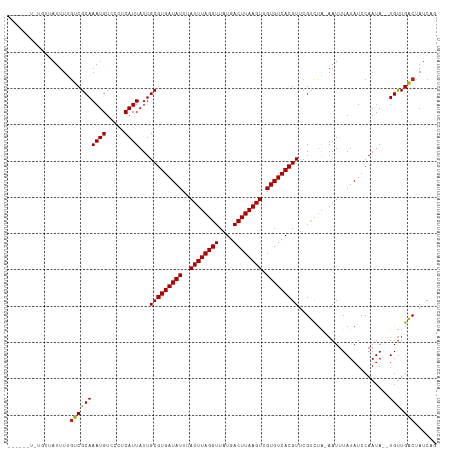
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:12:21 2011