| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,374,209 – 9,374,327 |
| Length | 118 |
| Max. P | 0.929719 |

| Location | 9,374,209 – 9,374,327 |
|---|---|
| Length | 118 |
| Sequences | 12 |
| Columns | 129 |
| Reading direction | reverse |
| Mean pairwise identity | 73.31 |
| Shannon entropy | 0.56316 |
| G+C content | 0.44176 |
| Mean single sequence MFE | -35.18 |
| Consensus MFE | -12.21 |
| Energy contribution | -12.33 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.929719 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
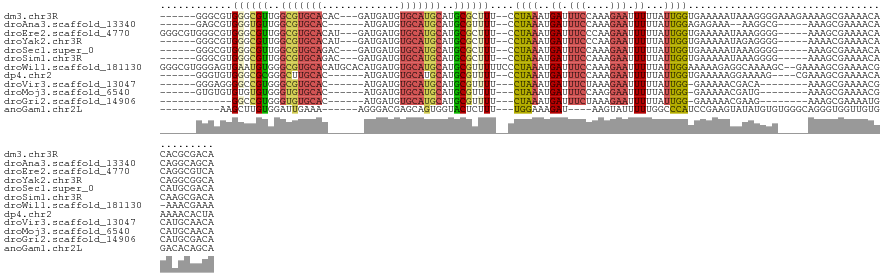
>dm3.chr3R 9374209 118 - 27905053 ------GGGCGUGGGCGUUGGCGUGCACAC---GAUGAUGUGCAUGCAUGCGCUUU--CCUAAAUGAUUUCCAAAGAAUUUUUAUUGGUGAAAAAUAAAGGGGAAAGAAAAGCGAAAACACACGCGACA ------..(((((...(((.(((((((((.---.....))))))))).(((.((((--(((.....((((((((.((....)).))))....))))....)))))))....)))..))).))))).... ( -37.00, z-score = -2.48, R) >droAna3.scaffold_13340 1926651 108 - 23697760 ------GAGCGUGGGUGUUGGCGUGCAC------AUGAUGUGCAUGCAUGCGUUUU--CCUAAAUGAUUUCCAAAGAAUUUUUAUUGGAGAGAAA--AAGGCG-----AAAGCGAAAACACAGGCAGCA ------..((((..(((((.((((((((------.....))))))))...((((((--(((......(((((((.((....)).)))))))....--..)).)-----))))))..)))))..)).)). ( -37.70, z-score = -3.40, R) >droEre2.scaffold_4770 5478186 119 + 17746568 GGGCGUGGGCGUGGGCGUUGGCGUGCACAU---GAUGAUGUGCAUGCAUGCGCUUU--CCUAAAUGAUUUCCCAAGAAUUUUUAUUGGUGAAAAAUAAAGGGG-----AAAGCGAAAACACAGGCGUCA .(((((..(.((..((((..((((((((((---....))))))))))))))(((((--(((......((((((((.........)))).)))).......)))-----)))))....)).)..))))). ( -45.82, z-score = -3.71, R) >droYak2.chr3R 13681165 113 - 28832112 ------GGGCGUGGGCGUUGGCGUGCACAU---GAUGAUGUGCAUGCAUGCGCUUU--CCUAAAUGAUUUCCCAAGAAUUUUUAUUGGUGAAAAAUAGAGGGG-----AAAACGAAAACACAGGCGGCA ------..((.(((((((..((((((((((---....))))))))))..)))))((--(((......((((((((.........)))).)))).......)))-----))..........)).)).... ( -38.42, z-score = -2.55, R) >droSec1.super_0 8487661 113 + 21120651 ------GGGCGUGGGCGUUGGCGUGCAGAC---GAUGAUGUGCAUGCAUGCGCUUU--CCUAAAUGAUUUCCAAAGAAUUUUUAUUGGUGAAAAAUAAAGGGG-----AAAGCGAAAACACAUGCGACA ------..(((((.((((..(((((((.(.---.....).)))))))))))(((((--(((.....((((((((.((....)).))))....))))....)))-----))))).......))))).... ( -35.10, z-score = -2.11, R) >droSim1.chr3R 15465778 113 + 27517382 ------GGGCGUGGGCGUUGGCGUGCAGAC---GAUGAUGUGCAUGCAUGCGCUUU--CCUAAAUGAUUUCCAAAGAAUUUUUAUUGGUGAAAAAUAAAGGGG-----AAAGCGAAAACACAAGCGACA ------..(((((.(((((..(((....))---)..))))).)))))...((((((--(((.....((((((((.((....)).))))....))))....)))-----))))))............... ( -32.40, z-score = -1.37, R) >droWil1.scaffold_181130 6445429 126 - 16660200 GGGCGUGGGAGUGAAUGUGGGCGUGCACAUGCACAUGAUGUGCAUGCAUGCGUUUUUCCCUAAAUGAUUUCCAAAGAAUUUUUAUUGGAAAAAGAGGCAAAAGC--GAAAAGCGAAAACG-AAACGAAA ..((..(((((.((((((..((((((((((.......))))))))))..))))))))))).......(((((((.((....)).))))))).....))....((--.....)).......-........ ( -37.20, z-score = -2.18, R) >dp4.chr2 16816723 111 - 30794189 ------GGGUGUGGGCGCGGGCUUGCAC------AUGAUGUGCAUGCAUGCGUUUU--CCUAAAUGAUUUCCAAAGAAUUUUUAUUGGUGAAAAAGGAAAAG----CGAAAGCGAAAACAAAAACACUA ------.(((((..(((((.((.(((((------.....))))).)).)))))(((--(((.........((((.((....)).))))......)))))).(----(....))..........))))). ( -32.76, z-score = -2.25, R) >droVir3.scaffold_13047 9162853 105 - 19223366 ------GGGAGGGGCCGUGGGCGUGCAC------AUGAUGUGCAUGCAUGCGUUUU---CUAAAUGAUUUCUAAAGAAUUUUUAUUGG-GAAAAACGACA--------AAAGCGAAAACGCAUGCAACA ------.......(((...)))((((((------.....))))))(((((((((((---(........(((....)))(((((.(((.-......))).)--------)))).)))))))))))).... ( -31.80, z-score = -2.42, R) >droMoj3.scaffold_6540 2670546 105 + 34148556 ------GUGUGUGUGUGUGGGUGUGCAC------AUGAUGUGCAUGCAUGCGUUUU---CUAAAUGAUUUCCAAGGAAUUUUUAUUGG-GAAAAACGAUG--------AAAGCGAAAACGCAUGCAACA ------(..(((((((((......))))------)).)))..).((((((((((((---(........(((....)))(((((((((.-......)))))--------)))).)))))))))))))... ( -36.70, z-score = -3.35, R) >droGri2.scaffold_14906 10090153 99 + 14172833 ------------GGCCGUGGGUGUGCAC------AUGAUGUGCAUGCAUGCGUUUU---CUAAAUGAUUUCUAAAGAAUUUUUAUUGG-GAAAAACGAAG--------AAAGCGAAAAUGCAUGCGACA ------------...((((........)------))).(((((((((((.((((((---((......(((((((.((....)).))))-)))......))--------))))))...)))))))).))) ( -29.30, z-score = -2.33, R) >anoGam1.chr2L 36203321 106 + 48795086 ----------AAGCUUGUGGAUUGAAA------AGGGACGAGCAGUGGUACUCUUU---UGGAAAGAU----AAGUAUUUUUGGCCCAUCCGAAGUAUAUGUGUGGGCAGGGUGGUUGUGGACACAGCA ----------..(((.(((..((.(((------((((((........)).))))))---).))...((----((.(((((...(((((..((.......))..))))).))))).))))...)))))). ( -28.00, z-score = -0.53, R) >consensus ______GGGCGUGGGCGUGGGCGUGCAC______AUGAUGUGCAUGCAUGCGCUUU__CCUAAAUGAUUUCCAAAGAAUUUUUAUUGGUGAAAAAUAAAGGGG_____AAAGCGAAAACACAUGCGACA ............((((((..(((((((.............)))))))..))))))....((((..((.(((....))).))...))))......................................... (-12.21 = -12.33 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:12:18 2011