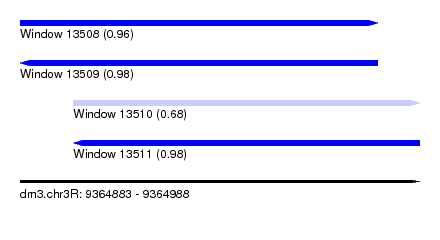
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,364,883 – 9,364,988 |
| Length | 105 |
| Max. P | 0.983445 |

| Location | 9,364,883 – 9,364,977 |
|---|---|
| Length | 94 |
| Sequences | 10 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 88.88 |
| Shannon entropy | 0.21157 |
| G+C content | 0.41903 |
| Mean single sequence MFE | -27.08 |
| Consensus MFE | -24.61 |
| Energy contribution | -24.73 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.961974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9364883 94 + 27905053 ---------------------CAAG-AGCGCGGCAUAGUAUGCCACACAAAUGCUGAAUAGUUGCGGAAGGUUAUGAAUUUCCGUGAGAAAUUACUACAAGUAAUAUGUGAGUGUU ---------------------....-((((((((((...))))).((((............(..((((((........))))))..)...(((((.....))))).)))).))))) ( -25.30, z-score = -2.23, R) >droSim1.chr3R 15456384 94 - 27517382 ---------------------CAAG-AGCGCGGCAUAGUAUGCCACACAAAUGCUGAAUAGUUGCGGAAGGUUAUGAAUUUCCGUGAGAAAUUACUACAAGUAAUAUGUGAGUGUU ---------------------....-((((((((((...))))).((((............(..((((((........))))))..)...(((((.....))))).)))).))))) ( -25.30, z-score = -2.23, R) >droSec1.super_0 8478368 94 - 21120651 ---------------------CAAG-AGCGCGGCAUAGUAUGCCACACAAAUGCUGAAUAGUUGCGGAAGGUUAUGAAUUUCCGUGAGAAAUUACUACAAGUAAUAUGUGAGUGUU ---------------------....-((((((((((...))))).((((............(..((((((........))))))..)...(((((.....))))).)))).))))) ( -25.30, z-score = -2.23, R) >droYak2.chr3R 13671434 94 + 28832112 ---------------------CAAG-AGCGCGGCAUAGUAUGCCACACAAAUGCUGAAUAGUUGCGGAAGGUUAUGAAUUUCCGUGAGAAAUUACUACAAGUAAUAUGUGAGUGUU ---------------------....-((((((((((...))))).((((............(..((((((........))))))..)...(((((.....))))).)))).))))) ( -25.30, z-score = -2.23, R) >droEre2.scaffold_4770 5468715 94 - 17746568 ---------------------CAAG-AGCGCGGCAUAGUAUGCCACACAAAUGCUGAAUAGUUGCGGAAGGUUAUGAAUUUCCGUGAGAAAUUACUACAAGUAAUAUGUGAGUGUU ---------------------....-((((((((((...))))).((((............(..((((((........))))))..)...(((((.....))))).)))).))))) ( -25.30, z-score = -2.23, R) >droAna3.scaffold_13340 1917570 94 + 23697760 ---------------------CAAG-AGCGCGGCAGAGUAUGCCACACAAAUGCUGAAUAGUUGCGGAAGGUUAUGAAUUUCCGUGAGAAAUUACUACAAGUAAUAUGUGAGUGUU ---------------------....-(((((((((.....)))).((((............(..((((((........))))))..)...(((((.....))))).)))).))))) ( -25.90, z-score = -2.53, R) >droPer1.super_0 8237238 115 - 11822988 GGCAGUGGCAGGGAGCUGUGGCAGG-AGCGCGGCAUAGUAUGCCACACAAAUGUUGAAUAGUUGCGGAAGGUUAUGAAUUUCCGUGAGAAAUUACUACAAGUAAUAUGUGAGUGUU ....((.((((....)))).))...-((((((((((...))))).((((............(..((((((........))))))..)...(((((.....))))).)))).))))) ( -34.10, z-score = -1.84, R) >dp4.chr2 16807315 115 + 30794189 GGCAGUGGCAGGGCGCUGUGGCAGG-AGCGCGGCAUAGUAUGCCACACAAAUGUUGAAUAGUUGCGGAAGGUUAUGAAUUUCCGUGAGAAAUUACUACAAGUAAUAUGUGAGUGUU ....((.((((....)))).))...-((((((((((...))))).((((............(..((((((........))))))..)...(((((.....))))).)))).))))) ( -32.60, z-score = -0.73, R) >droWil1.scaffold_181130 6435710 96 + 16660200 --------------------CUAGAACGCGCGGCAUAGUAUGCCACACAAAUGUUGAAUAGUUGCGGAAGGUUAUGAAUUUCCGUGAGAAAUUACUACAAGUAAUAUGUGAGUGGU --------------------......(((..(((((...))))).((((............(..((((((........))))))..)...(((((.....))))).)))).))).. ( -24.10, z-score = -1.55, R) >droVir3.scaffold_13047 9154564 103 + 19223366 -------------GAGAGUAGCAGCUAGCGCGGCAUAGUAUGCCACACAAAUGUUGAAUAGUUGCGGAAGGUUAUGAAUUUCCGUGAGAAAUUACUACAAGUAAUAUGUGAGUGCU -------------...(((....)))((((((((((...))))).((((............(..((((((........))))))..)...(((((.....))))).)))).))))) ( -27.60, z-score = -1.68, R) >consensus _____________________CAAG_AGCGCGGCAUAGUAUGCCACACAAAUGCUGAAUAGUUGCGGAAGGUUAUGAAUUUCCGUGAGAAAUUACUACAAGUAAUAUGUGAGUGUU ..........................(((((((((.....)))).((((............(..((((((........))))))..)...(((((.....))))).)))).))))) (-24.61 = -24.73 + 0.12)
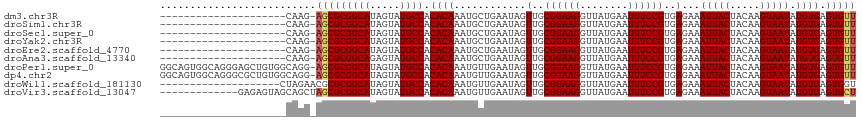
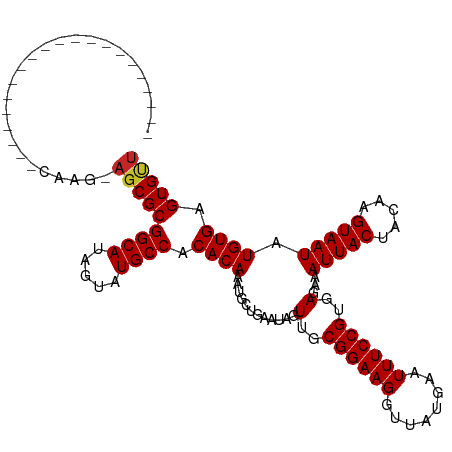
| Location | 9,364,883 – 9,364,977 |
|---|---|
| Length | 94 |
| Sequences | 10 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 88.88 |
| Shannon entropy | 0.21157 |
| G+C content | 0.41903 |
| Mean single sequence MFE | -19.67 |
| Consensus MFE | -18.25 |
| Energy contribution | -18.25 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.980112 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9364883 94 - 27905053 AACACUCACAUAUUACUUGUAGUAAUUUCUCACGGAAAUUCAUAACCUUCCGCAACUAUUCAGCAUUUGUGUGGCAUACUAUGCCGCGCU-CUUG--------------------- ...(((.(((.......)))))).........(((((..........)))))................(((((((((...))))))))).-....--------------------- ( -18.40, z-score = -2.10, R) >droSim1.chr3R 15456384 94 + 27517382 AACACUCACAUAUUACUUGUAGUAAUUUCUCACGGAAAUUCAUAACCUUCCGCAACUAUUCAGCAUUUGUGUGGCAUACUAUGCCGCGCU-CUUG--------------------- ...(((.(((.......)))))).........(((((..........)))))................(((((((((...))))))))).-....--------------------- ( -18.40, z-score = -2.10, R) >droSec1.super_0 8478368 94 + 21120651 AACACUCACAUAUUACUUGUAGUAAUUUCUCACGGAAAUUCAUAACCUUCCGCAACUAUUCAGCAUUUGUGUGGCAUACUAUGCCGCGCU-CUUG--------------------- ...(((.(((.......)))))).........(((((..........)))))................(((((((((...))))))))).-....--------------------- ( -18.40, z-score = -2.10, R) >droYak2.chr3R 13671434 94 - 28832112 AACACUCACAUAUUACUUGUAGUAAUUUCUCACGGAAAUUCAUAACCUUCCGCAACUAUUCAGCAUUUGUGUGGCAUACUAUGCCGCGCU-CUUG--------------------- ...(((.(((.......)))))).........(((((..........)))))................(((((((((...))))))))).-....--------------------- ( -18.40, z-score = -2.10, R) >droEre2.scaffold_4770 5468715 94 + 17746568 AACACUCACAUAUUACUUGUAGUAAUUUCUCACGGAAAUUCAUAACCUUCCGCAACUAUUCAGCAUUUGUGUGGCAUACUAUGCCGCGCU-CUUG--------------------- ...(((.(((.......)))))).........(((((..........)))))................(((((((((...))))))))).-....--------------------- ( -18.40, z-score = -2.10, R) >droAna3.scaffold_13340 1917570 94 - 23697760 AACACUCACAUAUUACUUGUAGUAAUUUCUCACGGAAAUUCAUAACCUUCCGCAACUAUUCAGCAUUUGUGUGGCAUACUCUGCCGCGCU-CUUG--------------------- ...(((.(((.......)))))).........(((((..........)))))................((((((((.....)))))))).-....--------------------- ( -18.20, z-score = -2.19, R) >droPer1.super_0 8237238 115 + 11822988 AACACUCACAUAUUACUUGUAGUAAUUUCUCACGGAAAUUCAUAACCUUCCGCAACUAUUCAACAUUUGUGUGGCAUACUAUGCCGCGCU-CCUGCCACAGCUCCCUGCCACUGCC ..................(((((..........((((..........))))(((..............(((((((((...))))))))).-...((....))....))).))))). ( -22.20, z-score = -1.86, R) >dp4.chr2 16807315 115 - 30794189 AACACUCACAUAUUACUUGUAGUAAUUUCUCACGGAAAUUCAUAACCUUCCGCAACUAUUCAACAUUUGUGUGGCAUACUAUGCCGCGCU-CCUGCCACAGCGCCCUGCCACUGCC ..................(((((..........((((..........))))(((..............(((((((((...))))))))).-...((....))....))).))))). ( -22.70, z-score = -1.43, R) >droWil1.scaffold_181130 6435710 96 - 16660200 ACCACUCACAUAUUACUUGUAGUAAUUUCUCACGGAAAUUCAUAACCUUCCGCAACUAUUCAACAUUUGUGUGGCAUACUAUGCCGCGCGUUCUAG-------------------- ...(((.(((.......)))))).........(((((..........)))))................(((((((((...))))))))).......-------------------- ( -18.90, z-score = -2.39, R) >droVir3.scaffold_13047 9154564 103 - 19223366 AGCACUCACAUAUUACUUGUAGUAAUUUCUCACGGAAAUUCAUAACCUUCCGCAACUAUUCAACAUUUGUGUGGCAUACUAUGCCGCGCUAGCUGCUACUCUC------------- ..................((((((........(((((..........)))))................(((((((((...)))))))))....))))))....------------- ( -22.70, z-score = -2.60, R) >consensus AACACUCACAUAUUACUUGUAGUAAUUUCUCACGGAAAUUCAUAACCUUCCGCAACUAUUCAGCAUUUGUGUGGCAUACUAUGCCGCGCU_CUUG_____________________ ...(((.(((.......)))))).........(((((..........)))))................((((((((.....))))))))........................... (-18.25 = -18.25 + 0.00)

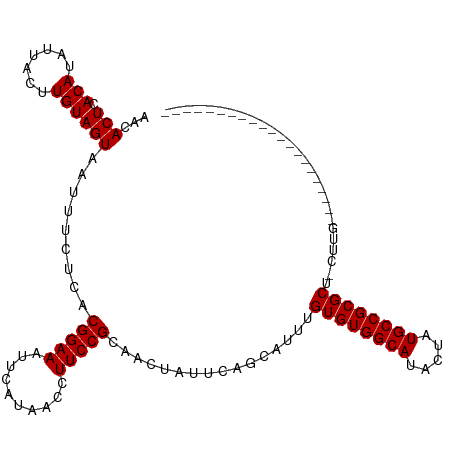
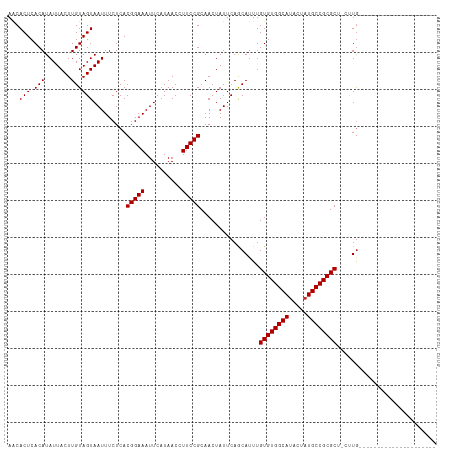
| Location | 9,364,897 – 9,364,988 |
|---|---|
| Length | 91 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.08 |
| Shannon entropy | 0.21045 |
| G+C content | 0.38142 |
| Mean single sequence MFE | -25.57 |
| Consensus MFE | -18.72 |
| Energy contribution | -18.79 |
| Covariance contribution | 0.07 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.677034 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9364897 91 + 27905053 AGUAUGCCACACAAAUGCUGAAUAGUUGCGGAAGGUUAUGAAUUUCCGUGAGAAAUUACUACAAGUAAUAUGUGAGUGUUGUAUGGGCAUG----------------------------- ..((((((.((..((((((.(...((((((((((........)))))((((....)))).....)))))...).))))))...))))))))----------------------------- ( -25.40, z-score = -2.51, R) >droSim1.chr3R 15456398 91 - 27517382 AGUAUGCCACACAAAUGCUGAAUAGUUGCGGAAGGUUAUGAAUUUCCGUGAGAAAUUACUACAAGUAAUAUGUGAGUGUUGUAUGGGCAUG----------------------------- ..((((((.((..((((((.(...((((((((((........)))))((((....)))).....)))))...).))))))...))))))))----------------------------- ( -25.40, z-score = -2.51, R) >droSec1.super_0 8478382 91 - 21120651 AGUAUGCCACACAAAUGCUGAAUAGUUGCGGAAGGUUAUGAAUUUCCGUGAGAAAUUACUACAAGUAAUAUGUGAGUGUUGUAUGGGCAUG----------------------------- ..((((((.((..((((((.(...((((((((((........)))))((((....)))).....)))))...).))))))...))))))))----------------------------- ( -25.40, z-score = -2.51, R) >droYak2.chr3R 13671448 91 + 28832112 AGUAUGCCACACAAAUGCUGAAUAGUUGCGGAAGGUUAUGAAUUUCCGUGAGAAAUUACUACAAGUAAUAUGUGAGUGUUGUAUGGGCAUG----------------------------- ..((((((.((..((((((.(...((((((((((........)))))((((....)))).....)))))...).))))))...))))))))----------------------------- ( -25.40, z-score = -2.51, R) >droEre2.scaffold_4770 5468729 91 - 17746568 AGUAUGCCACACAAAUGCUGAAUAGUUGCGGAAGGUUAUGAAUUUCCGUGAGAAAUUACUACAAGUAAUAUGUGAGUGUUGUAUGGGCAUG----------------------------- ..((((((.((..((((((.(...((((((((((........)))))((((....)))).....)))))...).))))))...))))))))----------------------------- ( -25.40, z-score = -2.51, R) >droAna3.scaffold_13340 1917584 91 + 23697760 AGUAUGCCACACAAAUGCUGAAUAGUUGCGGAAGGUUAUGAAUUUCCGUGAGAAAUUACUACAAGUAAUAUGUGAGUGUUAUAUGGGCAUG----------------------------- ..((((((.((..((((((.(...((((((((((........)))))((((....)))).....)))))...).))))))...))))))))----------------------------- ( -24.30, z-score = -2.34, R) >droPer1.super_0 8237273 91 - 11822988 AGUAUGCCACACAAAUGUUGAAUAGUUGCGGAAGGUUAUGAAUUUCCGUGAGAAAUUACUACAAGUAAUAUGUGAGUGUUAUAUGGGCAUG----------------------------- ..((((((.................(..((((((........))))))..)..........((.(((((((....))))))).))))))))----------------------------- ( -21.80, z-score = -1.67, R) >dp4.chr2 16807350 91 + 30794189 AGUAUGCCACACAAAUGUUGAAUAGUUGCGGAAGGUUAUGAAUUUCCGUGAGAAAUUACUACAAGUAAUAUGUGAGUGUUAUAUGGGCAUG----------------------------- ..((((((.................(..((((((........))))))..)..........((.(((((((....))))))).))))))))----------------------------- ( -21.80, z-score = -1.67, R) >droWil1.scaffold_181130 6435726 91 + 16660200 AGUAUGCCACACAAAUGUUGAAUAGUUGCGGAAGGUUAUGAAUUUCCGUGAGAAAUUACUACAAGUAAUAUGUGAGUGGUAUAUGGGCACA----------------------------- .(((((((((.((.((((((..((((.(((((((........)))))))((....))))))....)))))).)).))))))))).......----------------------------- ( -25.30, z-score = -2.60, R) >droVir3.scaffold_13047 9154587 120 + 19223366 AGUAUGCCACACAAAUGUUGAAUAGUUGCGGAAGGUUAUGAAUUUCCGUGAGAAAUUACUACAAGUAAUAUGUGAGUGCUAUAUGCGCAUACAUGUAUGUGCCGCCAGCGCGAUUAUGUG .........((((............(..((((((........))))))..)...(((((.....))))).)))).(((((....(((((((....)))))))....)))))......... ( -31.40, z-score = -0.62, R) >droMoj3.scaffold_6540 2662242 120 - 34148556 AGUAUGCCACACAAAUGUUGAAUAGUUGCGGAAGGUUAUGAAUUUCCGUGAGAAAUUACUACAAGUAAUAUGUGAGUGCUAUAUGCGCAUACAUGUAUAUGCUGCCAGCGCGAUUAUGUG .........((((...((((....((((((((((........)))))((((....))))....(((((((((((.((((.....)))).)))))))...))))).)))).))))..)))) ( -29.70, z-score = -0.17, R) >consensus AGUAUGCCACACAAAUGCUGAAUAGUUGCGGAAGGUUAUGAAUUUCCGUGAGAAAUUACUACAAGUAAUAUGUGAGUGUUAUAUGGGCAUG_____________________________ ..((((((.((((............(..((((((........))))))..)...(((((.....))))).))))...........))))))............................. (-18.72 = -18.79 + 0.07)

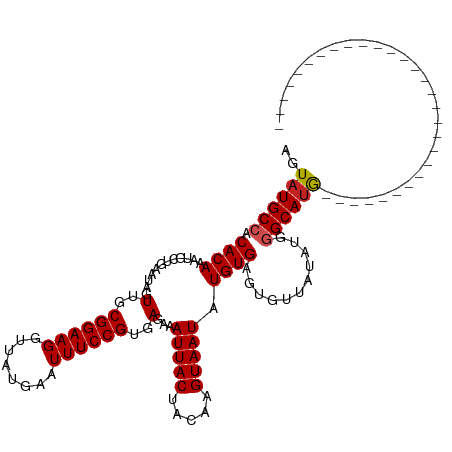
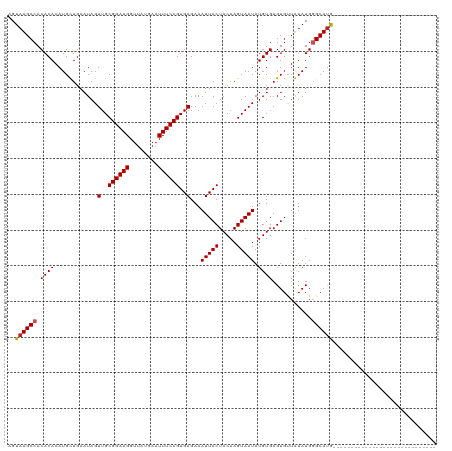
| Location | 9,364,897 – 9,364,988 |
|---|---|
| Length | 91 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.08 |
| Shannon entropy | 0.21045 |
| G+C content | 0.38142 |
| Mean single sequence MFE | -18.33 |
| Consensus MFE | -16.73 |
| Energy contribution | -16.36 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.983445 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9364897 91 - 27905053 -----------------------------CAUGCCCAUACAACACUCACAUAUUACUUGUAGUAAUUUCUCACGGAAAUUCAUAACCUUCCGCAACUAUUCAGCAUUUGUGUGGCAUACU -----------------------------.(((((.((((((.(((.(((.......)))))).........(((((..........)))))..............)))))))))))... ( -17.60, z-score = -2.74, R) >droSim1.chr3R 15456398 91 + 27517382 -----------------------------CAUGCCCAUACAACACUCACAUAUUACUUGUAGUAAUUUCUCACGGAAAUUCAUAACCUUCCGCAACUAUUCAGCAUUUGUGUGGCAUACU -----------------------------.(((((.((((((.(((.(((.......)))))).........(((((..........)))))..............)))))))))))... ( -17.60, z-score = -2.74, R) >droSec1.super_0 8478382 91 + 21120651 -----------------------------CAUGCCCAUACAACACUCACAUAUUACUUGUAGUAAUUUCUCACGGAAAUUCAUAACCUUCCGCAACUAUUCAGCAUUUGUGUGGCAUACU -----------------------------.(((((.((((((.(((.(((.......)))))).........(((((..........)))))..............)))))))))))... ( -17.60, z-score = -2.74, R) >droYak2.chr3R 13671448 91 - 28832112 -----------------------------CAUGCCCAUACAACACUCACAUAUUACUUGUAGUAAUUUCUCACGGAAAUUCAUAACCUUCCGCAACUAUUCAGCAUUUGUGUGGCAUACU -----------------------------.(((((.((((((.(((.(((.......)))))).........(((((..........)))))..............)))))))))))... ( -17.60, z-score = -2.74, R) >droEre2.scaffold_4770 5468729 91 + 17746568 -----------------------------CAUGCCCAUACAACACUCACAUAUUACUUGUAGUAAUUUCUCACGGAAAUUCAUAACCUUCCGCAACUAUUCAGCAUUUGUGUGGCAUACU -----------------------------.(((((.((((((.(((.(((.......)))))).........(((((..........)))))..............)))))))))))... ( -17.60, z-score = -2.74, R) >droAna3.scaffold_13340 1917584 91 - 23697760 -----------------------------CAUGCCCAUAUAACACUCACAUAUUACUUGUAGUAAUUUCUCACGGAAAUUCAUAACCUUCCGCAACUAUUCAGCAUUUGUGUGGCAUACU -----------------------------.(((((((((....(((.(((.......)))))).........(((((..........)))))...............)))).)))))... ( -15.80, z-score = -1.98, R) >droPer1.super_0 8237273 91 + 11822988 -----------------------------CAUGCCCAUAUAACACUCACAUAUUACUUGUAGUAAUUUCUCACGGAAAUUCAUAACCUUCCGCAACUAUUCAACAUUUGUGUGGCAUACU -----------------------------.(((((((((....(((.(((.......)))))).........(((((..........)))))...............)))).)))))... ( -15.80, z-score = -2.48, R) >dp4.chr2 16807350 91 - 30794189 -----------------------------CAUGCCCAUAUAACACUCACAUAUUACUUGUAGUAAUUUCUCACGGAAAUUCAUAACCUUCCGCAACUAUUCAACAUUUGUGUGGCAUACU -----------------------------.(((((((((....(((.(((.......)))))).........(((((..........)))))...............)))).)))))... ( -15.80, z-score = -2.48, R) >droWil1.scaffold_181130 6435726 91 - 16660200 -----------------------------UGUGCCCAUAUACCACUCACAUAUUACUUGUAGUAAUUUCUCACGGAAAUUCAUAACCUUCCGCAACUAUUCAACAUUUGUGUGGCAUACU -----------------------------((((((((((....(((.(((.......)))))).........(((((..........)))))...............)))).)))))).. ( -16.60, z-score = -2.16, R) >droVir3.scaffold_13047 9154587 120 - 19223366 CACAUAAUCGCGCUGGCGGCACAUACAUGUAUGCGCAUAUAGCACUCACAUAUUACUUGUAGUAAUUUCUCACGGAAAUUCAUAACCUUCCGCAACUAUUCAACAUUUGUGUGGCAUACU .........((((((...((.((((....)))).))...))))...((((((((((.....)))).......(((((..........)))))...............))))))))..... ( -25.40, z-score = -1.15, R) >droMoj3.scaffold_6540 2662242 120 + 34148556 CACAUAAUCGCGCUGGCAGCAUAUACAUGUAUGCGCAUAUAGCACUCACAUAUUACUUGUAGUAAUUUCUCACGGAAAUUCAUAACCUUCCGCAACUAUUCAACAUUUGUGUGGCAUACU .........((((((((.((((((....))))))))...))))...((((((((((.....)))).......(((((..........)))))...............))))))))..... ( -24.20, z-score = -1.00, R) >consensus _____________________________CAUGCCCAUAUAACACUCACAUAUUACUUGUAGUAAUUUCUCACGGAAAUUCAUAACCUUCCGCAACUAUUCAGCAUUUGUGUGGCAUACU ..............................((((.(((((((.(((.(((.......)))))).........(((((..........)))))..............)))))))))))... (-16.73 = -16.36 + -0.37)

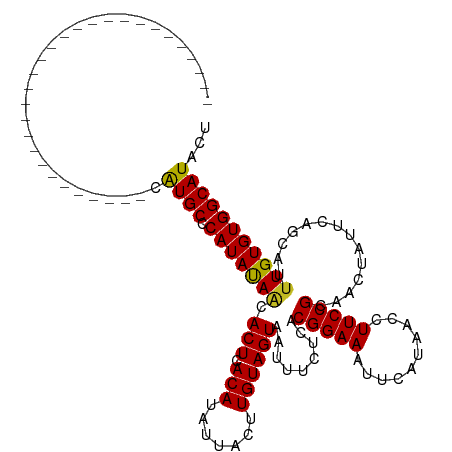
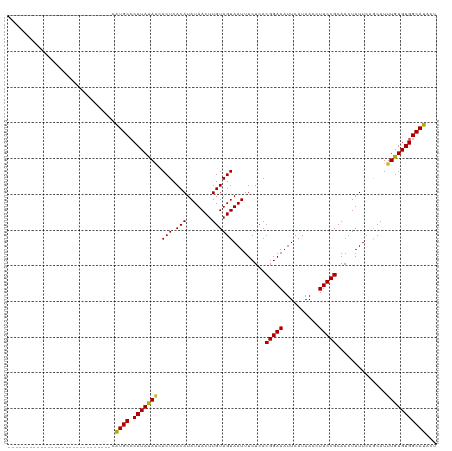
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:12:16 2011