| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,337,564 – 7,337,670 |
| Length | 106 |
| Max. P | 0.588202 |

| Location | 7,337,564 – 7,337,670 |
|---|---|
| Length | 106 |
| Sequences | 10 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 70.16 |
| Shannon entropy | 0.59346 |
| G+C content | 0.42421 |
| Mean single sequence MFE | -18.72 |
| Consensus MFE | -9.06 |
| Energy contribution | -9.18 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.33 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.588202 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
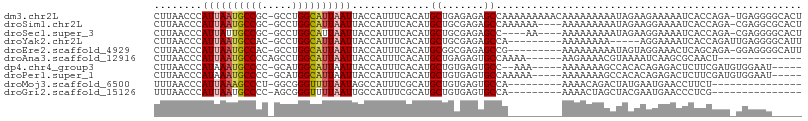
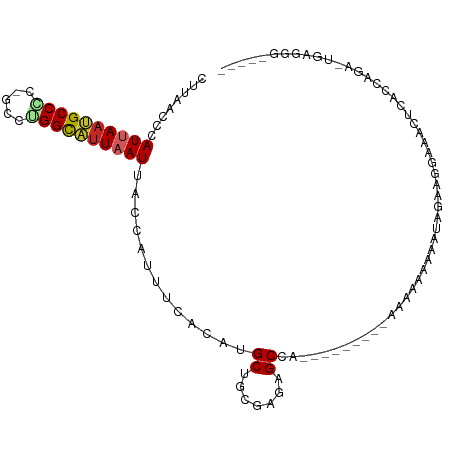
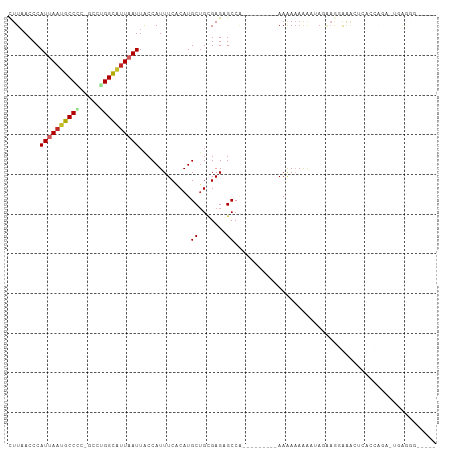
>dm3.chr2L 7337564 106 - 23011544 CUUAACCCAUUAAUGCCGC-GCCUGGCAUUAAUUACCAUUUCACAUGCUGAGAGAGCCAAAAAAAAACAAAAAAAAAUAGAAGAAAAAUCACCAGA-UGAGGGGCACU .....(((((((((((((.-...))))))))))..........(((.(((.(.((.................................)).)))))-)).)))..... ( -18.11, z-score = -0.89, R) >droSim1.chr2L 7132774 102 - 22036055 CUUAACCCAUUAAUGCCGC-GCCUGGCAUUAAUUACCAUUUCACAUGCUGCGAGAGCCAAAAAA----AAAAAAAAAUAGAAGGAAAAUCACCAGA-CGAGGCGCACU .................((-((((.((......................((....)).......----..............((.......)).).-).))))))... ( -17.70, z-score = -0.47, R) >droSec1.super_3 2859676 98 - 7220098 CUUAACCCAUUAUUGCCGC-GCCUGGCAUUAAUUACCAUUUCACAUGCUGCGAGAGCC----AA----AAAAAAAAAUAGAAGGAAAAUCACCAGA-CGAGGGGCACU .....(((((((.(((((.-...))))).)))).......((.......((....)).----..----..............((.......))...-.)))))..... ( -16.00, z-score = 0.69, R) >droYak2.chr2L 16763189 93 + 22324452 CUUAACCCAUUAAUGCCAC-GCCUGGCAUUAAUUACCAUUUCACAUGCUGCGAGAGCCA---------AAAAAAAA-----AGGAAAAUCACCAGAUUGAGGGGCAUU .....(((((((((((((.-...)))))))))).......(((....(((.(.((.((.---------........-----.))....)).))))..))))))..... ( -21.00, z-score = -1.31, R) >droEre2.scaffold_4929 16258960 97 - 26641161 CUUAACCCAUUAAUGCCAC-GCCUGGCAUUAAUUACCAUUUCACAUGCGGCGAGAGCCG---------AAAAAAAAAUAGUAGGAAACUCAGCAGA-GGAGGGGCAUU .....(((((((((((((.-...)))))))))).......((...((((((....))).---------...........(.((....))).)))..-.)))))..... ( -31.30, z-score = -3.74, R) >droAna3.scaffold_12916 15245359 88 - 16180835 CUUAACCCAUUAAUGCCCCAGCCUGGCAUUAAUUACCAUUUCACAUGCUGAGAGUGCCAAAA------AAGAAAACGUAAAAUCAAGCGCAACU-------------- ........(((((((((.......))))))))).....(((((.....)))))((((.....------..................))))....-------------- ( -13.30, z-score = -0.74, R) >dp4.chr4_group3 10946445 95 + 11692001 CUUAACCCAUAAAUGCCCC-GCAUGGCAUUAAUUACCAUUUCACAUGCUGUGAGUGCC--AAA-----AAAAAAAGCCACACAGAGACUCUUCGAUGUGGAAU----- ...........((((((..-....)))))).......(((((((((.(((((.(.((.--...-----.......))).))))).((....)).)))))))))----- ( -20.40, z-score = -1.81, R) >droPer1.super_1 8060759 97 + 10282868 CUUAACCCAUAAAUGCCCC-GCAUGGCAUUAAUUACCAUUUCACAUGCUGUGAGUGCCAAAAA-----AAAAAAAGCCACACAGAGACUCUUCGAUGUGGAAU----- ...........((((((..-....)))))).......(((((((((.(((((.(.((......-----.......))).))))).((....)).)))))))))----- ( -20.22, z-score = -1.79, R) >droMoj3.scaffold_6500 13831364 83 + 32352404 UUUAACCCAUUAAAGCCCU-GGCGGGUUUUAAUAGCCAUUUCGCAUGCUGUGAGUGCCA---------AAAACAGACUAUGAAUGAACCUUCU--------------- ........((((((((((.-...))))))))))...(((((......((((........---------...)))).....)))))........--------------- ( -16.40, z-score = -0.34, R) >droGri2.scaffold_15126 4913621 83 - 8399593 UUUAACCCAUUAAUGCCCC-AGCGGGUUUUAAUUGCCAUUUCGCAUGCUGUGAGUGCCA---------AAAACUAGCUACGAAUGAACCCUCG--------------- ........(((((.((((.-...)))).)))))...((((.((...(((...(((....---------...))))))..))))))........--------------- ( -12.80, z-score = 0.48, R) >consensus CUUAACCCAUUAAUGCCCC_GCCUGGCAUUAAUUACCAUUUCACAUGCUGCGAGAGCCA_________AAAAAAAAAUAGAAGGAAACUCACCAGA_UGAGGG_____ ........((((((((((.....)))))))))).............((.......))................................................... ( -9.06 = -9.18 + 0.12)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:23:24 2011