
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,357,967 – 9,358,075 |
| Length | 108 |
| Max. P | 0.730548 |

| Location | 9,357,967 – 9,358,070 |
|---|---|
| Length | 103 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 82.76 |
| Shannon entropy | 0.37469 |
| G+C content | 0.36489 |
| Mean single sequence MFE | -19.65 |
| Consensus MFE | -10.55 |
| Energy contribution | -10.53 |
| Covariance contribution | -0.01 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.730548 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
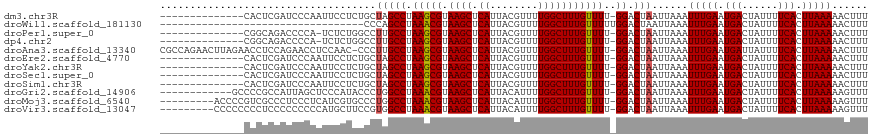
>dm3.chr3R 9357967 103 + 27905053 --------------CACUCGAUCCCAAUUCCUCUGCUAGCCUAAGCGUAAGCUCAUUACGUUUUGGCUUUGUUUU-GGACUAAUUAAAUUUGAAUGACUAUUUUCACUUAAAAACUUU --------------..............(((...((.((((.((((((((.....)))))))).))))..))...-))).........(((((.(((......))).)))))...... ( -21.40, z-score = -3.00, R) >droWil1.scaffold_181130 6426466 84 + 16660200 ----------------------------------CCCAGCCUAAACGUAAGCUCAUUACGUUUUGGCUUUGUUUUUGGACUAAUUAAAUUUGAAUGACUAUUUUCACUUAAAAACUUU ----------------------------------...((((.((((((((.....)))))))).))))....................(((((.(((......))).)))))...... ( -16.20, z-score = -2.56, R) >droPer1.super_0 8229515 102 - 11822988 --------------CGGCAGACCCCA-UCUCUGGCCUUGCCUAAGCGUAAGCUCAUUACGUUUUGGCUUUGUUUU-GGACUAAUUAAAUUUGAAUGACUAUUUUCACUUAAAAACUUU --------------.((((((.....-..))).)))..(((.((((((((.....)))))))).)))........-............(((((.(((......))).)))))...... ( -20.70, z-score = -1.18, R) >dp4.chr2 16799754 102 + 30794189 --------------CGGCAGACCCCA-UCUCUGGCCUUGCCUAAGCGUAAGCUCAUUACGUUUUGGCUUUGUUUU-GGACUAAUUAAAUUUGAAUGACUAUUUUCACUUAAAAACUUU --------------.((((((.....-..))).)))..(((.((((((((.....)))))))).)))........-............(((((.(((......))).)))))...... ( -20.70, z-score = -1.18, R) >droAna3.scaffold_13340 1910589 116 + 23697760 CGCCAGAACUUAGAACCUCCAGAACCUCCAAC-CCCUUGCCUAAGCGUAAGCUCAUUACGUUUUGGCUUUGUUUU-GGACUAAUUAAAUUUGAAUGAUUAUUUUCACUUAAAAACUUU .........((((....((((((((.......-.....(((.((((((((.....)))))))).)))...)))))-))))))).....(((((.(((......))).)))))...... ( -23.06, z-score = -2.62, R) >droEre2.scaffold_4770 5461624 103 - 17746568 --------------CACUCGAUCCCAAUUCCUCUGCUAGCCUAAGCGUAAGCUCAUUACGUUUUGGCUUUGUUUU-GGACUAAUUAAAUUUGAAUGACUAUUUUCACUUAAAAACUUU --------------..............(((...((.((((.((((((((.....)))))))).))))..))...-))).........(((((.(((......))).)))))...... ( -21.40, z-score = -3.00, R) >droYak2.chr3R 13664295 103 + 28832112 --------------CACUCGAUCCCAAUUCCUCUGCUAGCCUAAGCGUAAGCUCAUUACGUUUUGGCUUUGUUUU-GGACUAAUUAAAUUUGAAUGACUAUUUUCACUUAAAAACUUU --------------..............(((...((.((((.((((((((.....)))))))).))))..))...-))).........(((((.(((......))).)))))...... ( -21.40, z-score = -3.00, R) >droSec1.super_0 8471800 103 - 21120651 --------------CACUCGAUCCCAAUUCCUCUGCUAGCCUAAGCGUAAGCUCAUUACGUUUUGGCUUUGUUUU-GGACUAAUUAAAUUUGAAUGACUAUUUUCACUUAAAAACUUU --------------..............(((...((.((((.((((((((.....)))))))).))))..))...-))).........(((((.(((......))).)))))...... ( -21.40, z-score = -3.00, R) >droSim1.chr3R 15449571 103 - 27517382 --------------CACUCGAUCCCAAUUCCUCUGCUAGCCUAAGCGUAAGCUCAUUACGUUUUGGCUUUGUUUU-GGACUAAUUAAAUUUGAAUGACUAUUUUCACUUAAAAACUUU --------------..............(((...((.((((.((((((((.....)))))))).))))..))...-))).........(((((.(((......))).)))))...... ( -21.40, z-score = -3.00, R) >droGri2.scaffold_14906 10073449 105 - 14172833 ------------GCCCCGCCAUUAGCUCCCAUACCCUGGCCUAAACGUAAGCUCAUUACAUUUUGGCUUUGUUUU-GGACUAAUUAAAUUUGAAUGACUAUUUUCACUUAAAAAGUUU ------------........(((((.(((...((...((((.(((.((((.....)))).))).))))..))...-))))))))....(((((.(((......))).)))))...... ( -17.90, z-score = -0.96, R) >droMoj3.scaffold_6540 2653631 108 - 34148556 ---------ACCCCGUCGCCCUCCCUCAUCGUGCCCUGGCCUAAACGUAAGCUCAUUACAUUUUGGCUUUGUUUU-GGACUAAUUAAAUUUGAAUGACUAUUUUCACUUAAAAAGUUU ---------.....((((((....(.......)....)))(.(((((.((((.((........))))))))))).-))))........(((((.(((......))).)))))...... ( -14.00, z-score = 0.62, R) >droVir3.scaffold_13047 9147788 108 + 19223366 ---------CCCCCCCCUCCCCCCCCCAUGCUUCCGUGGCCUAAACGUAAGCUCAUUACAUUUUGGCUUUGUUUU-GGACUAAUUAAAUUUGAAUGACUAUUUUCACUUAAAAAGUUU ---------................(((((....)))))((.(((((.((((.((........))))))))))).-))..........(((((.(((......))).)))))...... ( -16.20, z-score = -0.96, R) >consensus ______________CACUCGAUCCCAAUUCCUCUCCUAGCCUAAGCGUAAGCUCAUUACGUUUUGGCUUUGUUUU_GGACUAAUUAAAUUUGAAUGACUAUUUUCACUUAAAAACUUU ....................................(((((..((((.((((.((........))))))))))...)).)))......(((((.(((......))).)))))...... (-10.55 = -10.53 + -0.01)

| Location | 9,357,973 – 9,358,075 |
|---|---|
| Length | 102 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 85.24 |
| Shannon entropy | 0.31796 |
| G+C content | 0.35274 |
| Mean single sequence MFE | -19.55 |
| Consensus MFE | -10.55 |
| Energy contribution | -10.53 |
| Covariance contribution | -0.01 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.596506 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9357973 102 + 27905053 -------------AUCCCAAUUCCUCUGCUAGCCUAAGCGUAAGCUCAUUACGUUUUGGCUUUGUUUU-GGACUAAUUAAAUUUGAAUGACUAUUUUCACUUAAAAACUUUUUGCC -------------........(((...((.((((.((((((((.....)))))))).))))..))...-))).........(((((.(((......))).)))))........... ( -21.40, z-score = -2.92, R) >droWil1.scaffold_181130 6426467 88 + 16660200 ----------------------------CCAGCCUAAACGUAAGCUCAUUACGUUUUGGCUUUGUUUUUGGACUAAUUAAAUUUGAAUGACUAUUUUCACUUAAAAACUUUUUGCC ----------------------------..((((.((((((((.....)))))))).))))....................(((((.(((......))).)))))........... ( -16.20, z-score = -2.05, R) >droPer1.super_0 8229521 101 - 11822988 --------------ACCCCAUCUCUGGCCUUGCCUAAGCGUAAGCUCAUUACGUUUUGGCUUUGUUUU-GGACUAAUUAAAUUUGAAUGACUAUUUUCACUUAAAAACUUUUUGCC --------------......(((..(((...(((.((((((((.....)))))))).)))...)))..-))).........(((((.(((......))).)))))........... ( -19.10, z-score = -1.80, R) >dp4.chr2 16799760 101 + 30794189 --------------ACCCCAUCUCUGGCCUUGCCUAAGCGUAAGCUCAUUACGUUUUGGCUUUGUUUU-GGACUAAUUAAAUUUGAAUGACUAUUUUCACUUAAAAACUUUUUGCC --------------......(((..(((...(((.((((((((.....)))))))).)))...)))..-))).........(((((.(((......))).)))))........... ( -19.10, z-score = -1.80, R) >droAna3.scaffold_13340 1910595 115 + 23697760 AACUUAGAACCUCCAGAACCUCCAACCCCUUGCCUAAGCGUAAGCUCAUUACGUUUUGGCUUUGUUUU-GGACUAAUUAAAUUUGAAUGAUUAUUUUCACUUAAAAACUUUUUGCC .............(((((..(((((..(...(((.((((((((.....)))))))).)))...)..))-))).........(((((.(((......))).)))))....))))).. ( -23.20, z-score = -3.02, R) >droEre2.scaffold_4770 5461630 102 - 17746568 -------------AUCCCAAUUCCUCUGCUAGCCUAAGCGUAAGCUCAUUACGUUUUGGCUUUGUUUU-GGACUAAUUAAAUUUGAAUGACUAUUUUCACUUAAAAACUUUUUGCC -------------........(((...((.((((.((((((((.....)))))))).))))..))...-))).........(((((.(((......))).)))))........... ( -21.40, z-score = -2.92, R) >droYak2.chr3R 13664301 102 + 28832112 -------------AUCCCAAUUCCUCUGCUAGCCUAAGCGUAAGCUCAUUACGUUUUGGCUUUGUUUU-GGACUAAUUAAAUUUGAAUGACUAUUUUCACUUAAAAACUUUUUGCC -------------........(((...((.((((.((((((((.....)))))))).))))..))...-))).........(((((.(((......))).)))))........... ( -21.40, z-score = -2.92, R) >droSec1.super_0 8471806 102 - 21120651 -------------AUCCCAAUUCCUCUGCUAGCCUAAGCGUAAGCUCAUUACGUUUUGGCUUUGUUUU-GGACUAAUUAAAUUUGAAUGACUAUUUUCACUUAAAAACUUUUUGCC -------------........(((...((.((((.((((((((.....)))))))).))))..))...-))).........(((((.(((......))).)))))........... ( -21.40, z-score = -2.92, R) >droSim1.chr3R 15449577 102 - 27517382 -------------AUCCCAAUUCCUCUGCUAGCCUAAGCGUAAGCUCAUUACGUUUUGGCUUUGUUUU-GGACUAAUUAAAUUUGAAUGACUAUUUUCACUUAAAAACUUUUUGCC -------------........(((...((.((((.((((((((.....)))))))).))))..))...-))).........(((((.(((......))).)))))........... ( -21.40, z-score = -2.92, R) >droGri2.scaffold_14906 10073455 103 - 14172833 -----------CCAUUAGCUCCCAUACCCUGGCCUAAACGUAAGCUCAUUACAUUUUGGCUUUGUUUU-GGACUAAUUAAAUUUGAAUGACUAUUUUCACUUAAAAAGUUUUCAC- -----------..(((((.(((...((...((((.(((.((((.....)))).))).))))..))...-))))))))......((((.((((.(((......))).)))))))).- ( -19.10, z-score = -2.07, R) >droMoj3.scaffold_6540 2653637 107 - 34148556 --------UCGCCCUCCCUCAUCGUGCCCUGGCCUAAACGUAAGCUCAUUACAUUUUGGCUUUGUUUU-GGACUAAUUAAAUUUGAAUGACUAUUUUCACUUAAAAAGUUUUUGCC --------..((.................(((((.(((((.((((.((........))))))))))).-)).)))......(((((.(((......))).)))))........)). ( -14.70, z-score = 0.37, R) >droVir3.scaffold_13047 9147794 106 + 19223366 --------CCUCCCCCCCCCAUGCUUCCGUGGCCUAAACGUAAGCUCAUUACAUUUUGGCUUUGUUUU-GGACUAAUUAAAUUUGAAUGACUAUUUUCACUUAAAAAGUUUUUGC- --------..........(((((....)))))((.(((((.((((.((........))))))))))).-))..........(((((.(((......))).)))))..........- ( -16.20, z-score = -0.42, R) >consensus _____________AUCCCAAUUCCUCUCCUAGCCUAAGCGUAAGCUCAUUACGUUUUGGCUUUGUUUU_GGACUAAUUAAAUUUGAAUGACUAUUUUCACUUAAAAACUUUUUGCC .............................(((((..((((.((((.((........))))))))))...)).)))......(((((.(((......))).)))))........... (-10.55 = -10.53 + -0.01)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:12:11 2011