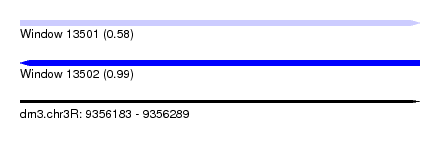
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,356,183 – 9,356,289 |
| Length | 106 |
| Max. P | 0.991840 |

| Location | 9,356,183 – 9,356,289 |
|---|---|
| Length | 106 |
| Sequences | 7 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 78.46 |
| Shannon entropy | 0.43854 |
| G+C content | 0.44666 |
| Mean single sequence MFE | -26.33 |
| Consensus MFE | -15.82 |
| Energy contribution | -17.80 |
| Covariance contribution | 1.98 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.577779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9356183 106 + 27905053 GACUCAGGAGUCUGUGGAUCCGGG-AGUUUGGGGUUCCGGGAGCAGCAGCAACAGCAAAACUAUUUCACAGCUAAAAAUGCUGUUAUGCGUUUAUAUUAACACGGAU .........(((((((...(((((-(.(....).))))))((((.(((..(((((((.....................))))))).))))))).......))))))) ( -30.00, z-score = -1.54, R) >droSim1.chr3R 15447817 105 - 27517382 GACUCAGGAGUCUGGGAAUCCGGG-AGUCUGGGGUUCCGGGAGCAGCAGCAACAGCAAAACUAUUUCACAGCUAAAAAUGCUGUUAUGCGUUUAUAU-AACACGGAU ..((((((..(((((....)))))-..))))))..((((......(((..(((((((.....................))))))).)))(((.....-))).)))). ( -31.70, z-score = -2.17, R) >droSec1.super_0 8470038 106 - 21120651 GACUCAGGAGUCUGGGAAUCCGGG-AGUCUGGGGUUCCGGGAGCAGCAGCAACAGCAAAACUAUUUCACAGCUAAAAAUGCUGUUAUGCGUUUAUAUUAACACGGAU ..((((((..(((((....)))))-..))))))..((((......(((..(((((((.....................))))))).)))(((......))).)))). ( -31.90, z-score = -2.17, R) >droYak2.chr3R 13662516 107 + 28832112 GGCCCGGGAGUCUGGGAAUUCUGGUAGUUCUGGGUUCCGGGAGCAGCAGCAACAGCAAAACUAUUUCAUAGCUAAAAAUGCUGUUAUGCGUUUAUAUUAACACGGAU .((((((((.((..(.....)..).).))))))))((((......(((..(((((((...((((...)))).......))))))).)))(((......))).)))). ( -31.50, z-score = -1.40, R) >droEre2.scaffold_4770 5459907 107 - 17746568 GGCUCGGGAGUCUGGGAGUCUGGGAAUUCUGGGGUUCCGGGAGCAGCAGCAACAGCAAAACUAUUUCACAGCUAAAAAUGCUGUUAUGCGUUUAUAUUAACACGGAU .((((.(((..((.(((((......))))).))..)))..)))).(((..(((((((.....................))))))).)))(((......)))...... ( -30.60, z-score = -1.48, R) >droAna3.scaffold_13340 1909320 80 + 23697760 ---------------------------CAUAGGAUGGGUUUUUUGGGAGCAACAGCAAAACUAUUUCACAGCUAAAAAUGCUGUUAUGCGUUUAUAUUAACACGGAU ---------------------------(((((((..(((((((((.......))).))))))..)))(((((.......))))))))).(((......)))...... ( -13.10, z-score = -0.27, R) >droWil1.scaffold_181130 6425101 93 + 16660200 -------------GUACAAUCAGGGUAAGUAGGG-GGAGUUGGGGGCAGCAACAGCAAAACUAUUUCACAGCUAAAAAUGCUGUUAUGCGUUUAUAUUAACACGGAU -------------.....(((...((.(((((..-(.((((....((.......))..)))).)..)(((((.......)))))..........)))).))...))) ( -15.50, z-score = 0.19, R) >consensus GACUCAGGAGUCUGGGAAUCCGGG_AGUCUGGGGUUCCGGGAGCAGCAGCAACAGCAAAACUAUUUCACAGCUAAAAAUGCUGUUAUGCGUUUAUAUUAACACGGAU ..........(((.(((((((.((....)).))))))).)))...(((..(((((((.....................))))))).)))(((......)))...... (-15.82 = -17.80 + 1.98)

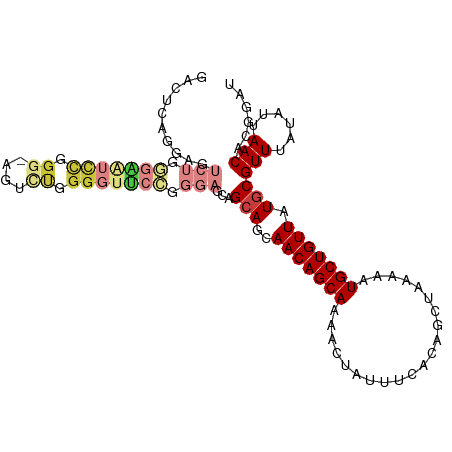
| Location | 9,356,183 – 9,356,289 |
|---|---|
| Length | 106 |
| Sequences | 7 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 78.46 |
| Shannon entropy | 0.43854 |
| G+C content | 0.44666 |
| Mean single sequence MFE | -22.79 |
| Consensus MFE | -15.91 |
| Energy contribution | -16.93 |
| Covariance contribution | 1.02 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.991840 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9356183 106 - 27905053 AUCCGUGUUAAUAUAAACGCAUAACAGCAUUUUUAGCUGUGAAAUAGUUUUGCUGUUGCUGCUGCUCCCGGAACCCCAAACU-CCCGGAUCCACAGACUCCUGAGUC ....(((...........(((((((((((.....((((((...)))))).)))))))).)))...(((.(((.........)-)).)))..))).((((....)))) ( -26.20, z-score = -2.35, R) >droSim1.chr3R 15447817 105 + 27517382 AUCCGUGUU-AUAUAAACGCAUAACAGCAUUUUUAGCUGUGAAAUAGUUUUGCUGUUGCUGCUGCUCCCGGAACCCCAGACU-CCCGGAUUCCCAGACUCCUGAGUC .((((.(((-.....)))(((((((((((.....((((((...)))))).)))))))).)))......))))......((((-(..(((.((...)).))).))))) ( -28.30, z-score = -2.90, R) >droSec1.super_0 8470038 106 + 21120651 AUCCGUGUUAAUAUAAACGCAUAACAGCAUUUUUAGCUGUGAAAUAGUUUUGCUGUUGCUGCUGCUCCCGGAACCCCAGACU-CCCGGAUUCCCAGACUCCUGAGUC .((((.(((......)))(((((((((((.....((((((...)))))).)))))))).)))......))))......((((-(..(((.((...)).))).))))) ( -28.50, z-score = -2.97, R) >droYak2.chr3R 13662516 107 - 28832112 AUCCGUGUUAAUAUAAACGCAUAACAGCAUUUUUAGCUAUGAAAUAGUUUUGCUGUUGCUGCUGCUCCCGGAACCCAGAACUACCAGAAUUCCCAGACUCCCGGGCC .((((.(((......)))(((((((((((.....((((((...)))))).)))))))).)))......))))...................(((........))).. ( -23.70, z-score = -1.19, R) >droEre2.scaffold_4770 5459907 107 + 17746568 AUCCGUGUUAAUAUAAACGCAUAACAGCAUUUUUAGCUGUGAAAUAGUUUUGCUGUUGCUGCUGCUCCCGGAACCCCAGAAUUCCCAGACUCCCAGACUCCCGAGCC ..................(((((((((((.....((((((...)))))).)))))))).))).((((..((((........))))..((.((...)).))..)))). ( -21.60, z-score = -1.40, R) >droAna3.scaffold_13340 1909320 80 - 23697760 AUCCGUGUUAAUAUAAACGCAUAACAGCAUUUUUAGCUGUGAAAUAGUUUUGCUGUUGCUCCCAAAAAACCCAUCCUAUG--------------------------- ....(((((......))))).((((((((.....((((((...)))))).))))))))......................--------------------------- ( -15.60, z-score = -2.94, R) >droWil1.scaffold_181130 6425101 93 - 16660200 AUCCGUGUUAAUAUAAACGCAUAACAGCAUUUUUAGCUGUGAAAUAGUUUUGCUGUUGCUGCCCCCAACUCC-CCCUACUUACCCUGAUUGUAC------------- ....(((((......))))).((((((((.....((((((...)))))).))))))))..............-.....................------------- ( -15.60, z-score = -1.98, R) >consensus AUCCGUGUUAAUAUAAACGCAUAACAGCAUUUUUAGCUGUGAAAUAGUUUUGCUGUUGCUGCUGCUCCCGGAACCCCAGACU_CCCGGAUUCCCAGACUCCUGAGUC .((((.(((......)))(((((((((((.....((((((...)))))).)))))))).)))......))))................................... (-15.91 = -16.93 + 1.02)
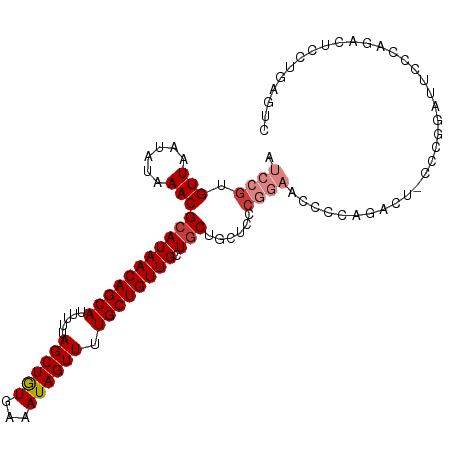
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:12:08 2011